

Editor | Scientists have been looking for efficient ways to predict the fit between these "keys" and "locks," or protein-ligand interactions.
However, traditional data-driven methods often fall into "rote learning", memorizing ligand and protein training data instead of truly learning the interactions between them.
Recently, a research team from Zhejiang University and the Chinese Academy of Sciences proposed a new scoring method called EquiScore, which uses heterogeneous graph neural networks to integrate physical prior knowledge and represent proteins in the equation transformation space - Ligand interactions.
EquiScore is trained on a new dataset built using multiple data augmentation strategies and a rigorous redundancy elimination scheme.
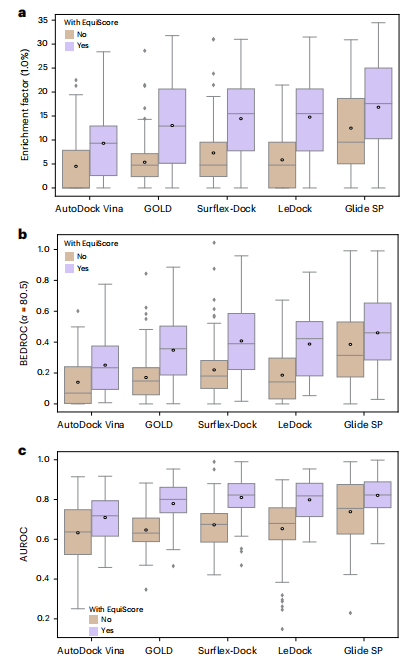
On two large external test sets, EquiScore started to come out on top compared to 21 other methods. When EquiScore is used with different docking methods, it can effectively enhance the screening capabilities of these docking methods. EquiScore also performed well in the task of ranking the activity of a series of structurally similar substances, demonstrating its potential to guide lead compound optimization.
Finally, different interpretability levels of EquiScore are studied, which may provide more insights for structure-based drug design.
The study is titled "
Generic protein–ligand interaction scoring by integrating physical prior knowledge and data augmentation modeling" and was published in "Nature Machine" on June 6, 2024 Intelligence》on.
 Paper link:
Paper link:
With the explosion of experimental protein-ligand interaction data, machine learning-based scoring methods have made substantial progress.
The increasing capacity of machine learning models enables them to remember the entire training data set. At the same time, data leakage issues between training data and test data lead to overly optimistic evaluations of the capabilities of these models
In addition to the quality of the data set, another key factor affecting the performance of machine learning-based scoring methods is Efficiently integrate physical prior information about ligand-protein interactions.
EquiScore's architectureFirst, the researchers built a new dataset called PDBscreen using multiple data augmentation strategies. For example, using close-to-native ligand binding poses to amplify the size of positive samples, and using generated highly deceptive decoys to amplify the size of negative samples.
Secondly, by introducing new types of nodes and edges and an information-aware attention mechanism, a heterogeneous graph that can integrate prior information on physical intermolecular interactions is proposed.
 Illustration: Pipeline for building PDBscreen dataset. (Source: Paper)
Illustration: Pipeline for building PDBscreen dataset. (Source: Paper)
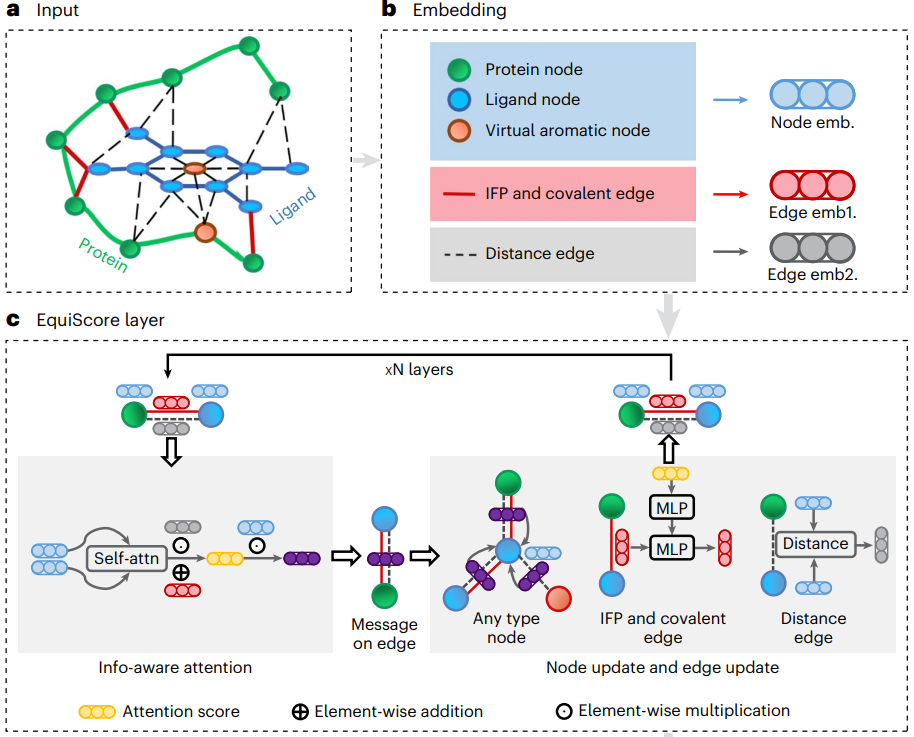
 Illustration: EquiScore overall architecture.
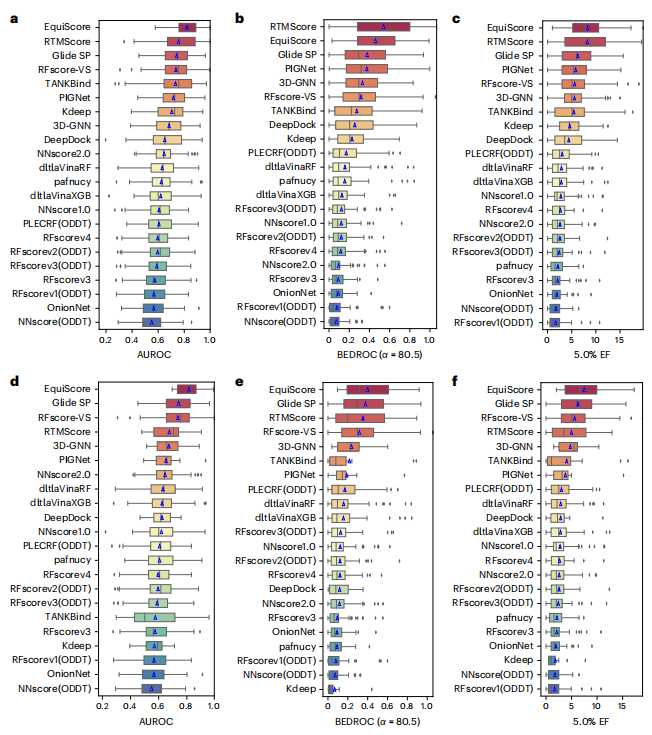
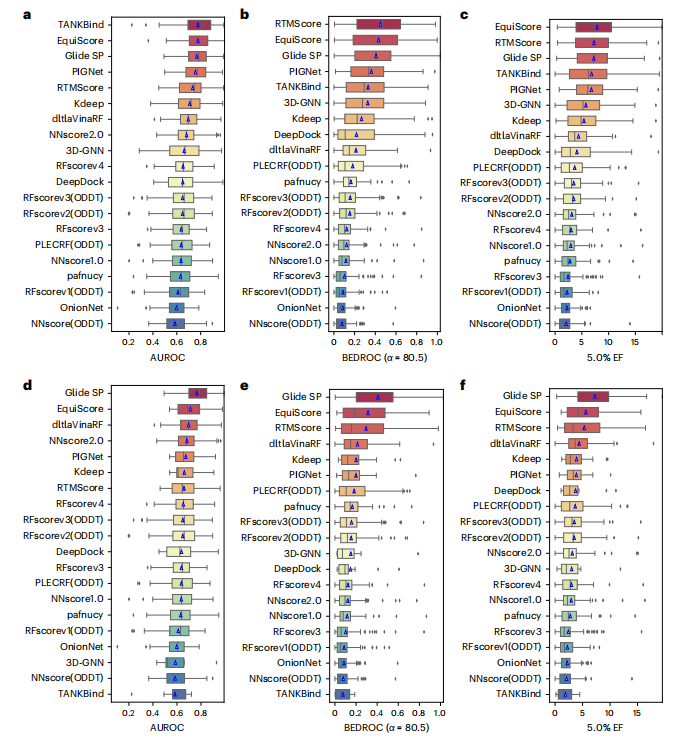
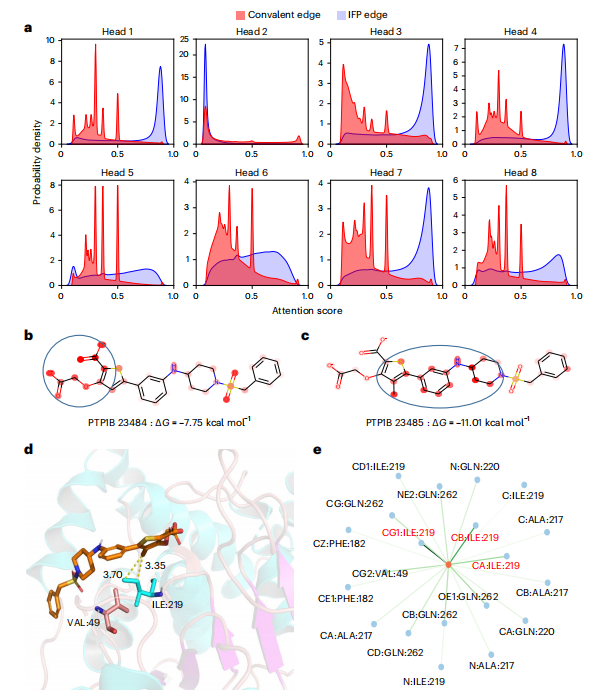
Illustration: EquiScore overall architecture. geometric) and structure-based edges through chemical bonds (Estructural) are established between nodes. The researchers also added a class of edges based on protein-ligand empirical interaction components (IFPs) calculated by ProLIF to Estructural to include a priori physical knowledge about intermolecular interactions. In the second step, an embedding layer is used to obtain a latent representation of each type of edges and nodes on the heterogeneous graph. This scheme can introduce other new nodes and edges with clear physical meaning, and can be seamlessly integrated with subsequent representation learning modules. In order to fully utilize the inductive bias of information from different nodes and edges while ensuring equal variance of the model, the EquiScore layer consists of three sub-modules: the information-aware attention module, the node update module and the edge update module. The information-aware attention module can interpret interactions from different information, including (1) equivariant geometric information, (2) chemical structure information, and (3) protein-ligand empirical interaction components. The researchers evaluated the performance of the generated EquiScore model. In the virtual screening (VS) scenario, EquiScore consistently achieved top rankings compared to 21 existing scoring methods for unseen proteins on two external datasets, DEKOIS2.0 and DUD-E. In the lead optimization scenario, EquiScore only showed lower ranking power compared to FEP+ among eight different methods. Considering that FEP+ calculations require significantly higher computational costs, EquiScore demonstrates a more balanced advantage between speed and accuracy. Furthermore, it was found that EquiScore exhibits strong rescoring capabilities when applied to poses generated by different docking methods, and that using EquiScore rescoring can improve VS performance for all evaluation methods. Finally, the researchers analyzed the interpretability of the model and found that the model could capture key intermolecular interactions, proving the rationality of the model and providing useful information for rational drug design. clues. Robust predictions of protein-ligand interactions will provide valuable opportunities to understand the biology of proteins and determine their impact on future drug therapies. EquiScore will contribute to a better understanding of human health and disease and facilitate the discovery of new drugs. Model Performance Evaluation
The above is the detailed content of 'AI+physics prior knowledge', Zhejiang University and Chinese Academy of Sciences general protein-ligand interaction scoring method published in Nature sub-journal. For more information, please follow other related articles on the PHP Chinese website!




