Technology peripherals
Technology peripherals
 AI
AI
 'Encyclopedia' of AI small molecule drug discovery, reviewed by researchers from Cornell, Cambridge, EPFL and others published in Nature sub-journal
'Encyclopedia' of AI small molecule drug discovery, reviewed by researchers from Cornell, Cambridge, EPFL and others published in Nature sub-journal
'Encyclopedia' of AI small molecule drug discovery, reviewed by researchers from Cornell, Cambridge, EPFL and others published in Nature sub-journal

Author | Cornell University Du Yuanqi
Editor | ScienceAI
As AI for Science receives more and more attention, people are more concerned about how AI can solve a series of scientific problems and can be successfully used for reference in other countries similar fields.
AI and small molecule drug discovery is one of the most representative and early explored fields. Molecular discovery is a very difficult combinatorial optimization problem (due to the discrete nature of the molecular structure) and the search space is very large and rugged. At the same time, it is very difficult to verify the properties of the searched molecules. It usually requires expensive experiments, at least simulation calculations, Quantum chemical methods to provide feedback.
With the rapid development of machine learning and thanks to early exploration (including the construction of simple and usable optimization goals and effect measurement methods), a large number of algorithms have been developed, including combinatorial optimization, search, sampling algorithms (genetic algorithms, Monte Carlo tree search, reinforcement learning, generative flow model/GFlowNet, Markov chain Monte Carlo, etc.), and continuous optimization algorithms, Bayesian optimization, gradient-based optimization, etc. At the same time, the existing relatively complete algorithm measurement benchmarks and relatively objective and fair comparison methods have also opened up a broad space for the development of machine learning algorithms.
Recently, researchers from Cornell University, University of Cambridge and Ecole Polytechnique Fédérale de Lausanne (EPFL) published a review article titled "Machine learning-aided generative molecular design" in "Nature Machine Intelligence".

Paper link: https://www.nature.com/articles/s42256-024-00843-5
This review reviews the application of machine learning in generative molecular design. Drug discovery and development requires optimizing molecules to meet specific physicochemical properties and biological activities. However, traditional methods are expensive and prone to failure due to the huge search space and discontinuous optimization functions. Machine learning accelerates the early-stage drug discovery process by combining molecule generation and screening steps.

Illustration: Generative ML assisted molecular design process.
Generative molecular design tasks
Generative molecular design can be divided into two major paradigms: distributed learning and goal-oriented generation, among which goal-oriented generation can be further divided into conditional generation and molecular optimization. The suitability of each method depends on the specific task and the data involved.
Distribution learning (distribution learning)
- Distribution learning aims to describe the distribution of data by modeling the probability distribution of molecules in a given data set, thereby sampling new molecules from the learned distribution.
Conditional generation (conditional generation)
- Property-conditioned generation: Generate a structure with specific attributes, which can be a text description or a value of a specific attribute.
- Molecular (sub)structure-conditioned generation(molecular (sub)structure-conditioned generation): Generate molecules with specific structural constraints, such as designing partial structures, scaffold hopping, linker design, redesigning the entire structure (pilot optimization) or the entire Conditional generation of molecules (conformational generation).
- Target-conditioned generation (target-conditioned generation): Aims to generate molecules with high binding affinity for specific disease-related biomolecular targets. Unlike attribute condition generation, target condition generation utilizes explicit access to the target structure to improve the affinity of the ligand molecule to the target by integrating direct target-ligand interactions.
- Phenotype-conditioned generation (phenotype-conditioned generation): Involves learning phenotypic fingerprints from cell-based microscopy or other bioassay readouts (such as transcriptome data) to provide conditioned signals that guide generation toward the desired biology The resulting numerator.
Molecular optimization plays a key role in drug discovery by refining the properties of drug candidates to improve their safety, efficacy and pharmacokinetic properties. Involves making small modifications to candidate molecular structures to optimize drug properties such as solubility, bioavailability, and target affinity, thereby improving therapeutic potential and increasing success with clinical endpoints.
-
Illustrations: Illustrations of generation tasks, generation strategies, and molecular characterization.
Molecular generation process
Molecular generation is a complex process including many different combination units. We list the representative work in the figure below and introduce the representative units of each part.
Molecular Representation
When developing molecularly generated neural architectures, it is first necessary to determine machine-readable input and output representations of the molecular structure. The input representation helps inject appropriate inductive biases into the model, while the output representation determines the optimized search space for the molecule. The representation type determines the applicability of the generation method, for example, discrete search algorithms can only be applied to combinatorial representations such as graphs and strings.
While various input representations have been studied, the trade-offs between representation types and the neural architectures that encode them are not yet clear. Representation transformations between molecules are not necessarily bijective; for example, density maps and fingerprints cannot uniquely identify molecules, and further techniques are needed to solve this non-trivial mapping problem. Common molecular representations include strings, two-dimensional topological graphs, and three-dimensional geometric graphs.
- String-based molecular structures: usually encoded as strings, such as Simplified Molecular Input Line Entry System (SMILES) or Self-Referential Embedded Strings (SELFIES). SMILES represents the molecule using syntax rules, but the string may be invalid; SELFIES determines the validity of the molecule by modifying these rules. Molecular strings are typically encoded into sequence data via recurrent networks and Transformer models.
- Atoms and bonds based on topological and geometric graphs: usually represented as nodes and edges in topological graphs. Graph neural networks (GNNs) are often used to model graph-structured molecular data, updating node and edge features based on adjacent nodes. Geometric GNNs are often used to capture application-relevant symmetries in 3D space, such as translation and rotation invariance or equivariance, when 3D information is available and relevant.
Representation granularity is another consideration in generative model design. Typically, methods utilize atoms or molecular fragments as basic building blocks during generation. Fragment-based representation refines molecular structures into larger units containing groups of atoms, carrying hierarchical information such as functional group identification, thereby aligning with traditional fragment-based or pharmacophore drug design approaches.
Generative methods
Deep generative models are a class of methods that estimate the probability distribution of data and sample from a learning distribution (also called distribution learning). These include variational autoencoders, generative adversarial networks, normalizing flows, autoregressive models, and diffusion models. Each of these generation methods has its use cases, pros and cons, and the choice depends on the required task and data characteristics.
Generation strategy
Generation strategy refers to the way the model outputs the molecular structure, which can generally be divided into one-time generation, sequential generation or iterative improvement.
One-shot generation: One-shot generation generates the complete molecular structure in a single forward pass of the model. This approach often struggles to generate realistic and reasonable molecular structures with high accuracy. Furthermore, one-shot generation often cannot satisfy explicit constraints, such as valence constraints, which are crucial to ensure the accuracy and validity of the generated structure.
Sequential Generation: Sequential generation builds a molecular structure through a series of steps, usually by atoms or fragments. Valence constraints can be easily injected into sequential generation, thereby improving the quality of the generated molecules. However, the main limitation of sequential generation is that the order of generated trajectories needs to be defined during training and is slower in inference.
Iterative improvement: Iterative improvement adjusts the prediction by predicting a series of updates, circumventing the difficulties in one-shot generation methods. For example, the cyclic structure module in AlphaFold2 successfully refined the backbone framework, an approach that inspired related molecule generation strategies. Diffusion modeling is a common technique that generates new data through a series of noise reduction steps. Currently, diffusion models have been applied to a variety of molecule generation problems, including conformational generation, structure-based drug design, and linker design.
Optimization strategy
Combination optimization: For the combinatorial encoding of molecules (pictures or strings), techniques in the field of combinatorial optimization can be directly applied.
Continuous Optimization: Molecules can be represented or encoded in continuous domains, such as point clouds and geometric maps in Euclidean space, or deep generative models that encode discrete data in continuous latent space.

Evaluation of Generative Machine Learning Models
Evaluating generative models requires computational evaluation and experimental verification. Standard metrics include effectiveness, uniqueness, novelty, etc. Multiple metrics should be considered when evaluating a model to fully assess build performance.
Experimental verification
產生的分子必須透過濕法實驗來進行明確的驗證,這與現有研究主要關注計算貢獻形成鮮明對比。雖然生成模型並非沒有弱點,但預測與實驗之間的脫節也歸因於進行此類驗證所需的專業知識、昂貴的費用、以及漫長的測試週期。


產生模型規律
大多數報告實驗驗證的研究使用 RNN 和/或 VAE,並以 SMILES 作為操作對象。我們總結了四個主要觀察點:
- SMILES 雖然捕捉到的 3D 資訊有限,但作為一種高效的表示方式,適用於分佈學習和小資料集的微調。
- 許多實驗驗證的研究目標是激酶,這是 ChEMBL 等流行開源資料集中的常見目標。
- 絕大多數目標導向的方法使用強化學習(單獨或作為組件)作為最佳化演算法,包括基於配體和基於結構的藥物設計。
- AlphaFold 預測的結構可以成功用於產生結構的藥物設計。
未來方向
儘管機器學習演算法為小分子藥物發現帶來了曙光,但是還有更多的挑戰與機遇需要面對。
挑戰
- 分佈外生成:已知化學物質只佔化學空間的一小部分。雖然深度生成模型可以提出訓練分佈之外的分子,但需要確保其合理性。
- 不現實的問題表述:精確的問題表述對於開發適用於現實世界藥物發現的模型至關重要。常常忽略的基本面向包括構象動態、水的作用和熵貢獻,而諸如無限訪問 oracle 呼叫的假設也常被錯誤地認為是理所當然的。這包含了樣本效率問題,最近的研究在有限 oracle 預算下的高效目標導向生成方面取得了進展。
- 低保真 oracle:在藥物發現相關維度上有效評分設計仍然困難,成為工業環境中部署生成模型的瓶頸。例如,高通量結合親和力預測在數據驅動和基於物理的工作流程中通常不準確。雖然存在替代的高精度 oracle,但其運算需求限制了可擴展性。此外,高品質標註資料的不可取得性也成為開發具有高精度和可管理 AI oracle 的障礙。
- 缺乏統一的評估協議:用於評估藥物候選物品質的評估協議與我們定義何為良好藥物的標準密切相關。 ML 社群通常使用的易於計算的物理化學描述符存在疑問,肯定無法全面反映性能。在生成分子設計與虛擬篩選之間進行嚴格比較也較少見。
- 缺乏大規模研究和基準測試:許多ML方法已經開發出來,但在許多關鍵任務中的不同模型類型上沒有公平的基準測試結果。例如,僅使用了可用資料的一小部分進行訓練,限制了對模型可擴展性的理解。最近的基準測試對標準化計算評估協議的重要貢獻。
- 缺乏可解釋性:可解釋性是分子生成模型中一個重要但未充分探索的領域。例如,洞察生成或優化過程如何建構分子可以產生化學規則,這對藥物化學家具有解釋性。這在小分子領域尤其重要,因為生成模型通常用於向藥物化學家提交想法,合成障礙排除了測試所有生成設計的可能性。
機會
- 超越小分子设计的应用:这里讨论的方法可能在设计其他复杂结构材料(如多糖、蛋白质(特别是抗体)、核酸、晶体结构和聚合物)方面有更广泛的应用。
- 大语言模型展示了通过文本指导的发现和决策作为代理来革新分子设计的潜力,这得益于大量可用的训练数据,包括科学文献。此外,针对分子结构进行定制或微调的模型为研究人员提供了利用自然语言处理中的成熟进展的额外机会。
- 药物开发的后期阶段:分子设计/优化占据了药物发现的早期阶段。然而,由于有限的疗效、较差的 ADME/T(吸收、分布、新陈代谢、排泄和毒性)特性和安全问题导致的晚期失败是药物开发管道中的痛点。尽管有限,但将临床数据集成到设计管道中是提高下游成功率的一个有希望的方向。
- 聚焦模型目的:药物发现管道是制药公司多年经验和艰难教训的结果。ML 研究人员应该不仅仅设计纯粹的从头设计模型(特别是在缺乏深度表征能力时),还应设计聚焦于在多年过程中的特定步骤上改进的模型,符合现实约束。
- 自动化实验室:对高通量实验的需求不断增加,以为 ML 设计的分子提供反馈,将越来越多的注意力集中在自动化实验室上,以加快设计–制造–测试–分析循环。
作者: 杜沅岂,康奈尔大学计算机系二年级博士生,主要研究兴趣,几何深度学习,概率模型,采样,搜索,优化问题,可解释性,与在分子探索领域的应用,具体信息见:https://yuanqidu.github.io/。
The above is the detailed content of 'Encyclopedia' of AI small molecule drug discovery, reviewed by researchers from Cornell, Cambridge, EPFL and others published in Nature sub-journal. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

AI Hentai Generator
Generate AI Hentai for free.

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1382
1382
 52
52
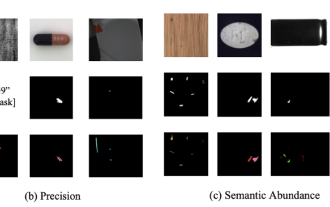 Breaking through the boundaries of traditional defect detection, 'Defect Spectrum' achieves ultra-high-precision and rich semantic industrial defect detection for the first time.
Jul 26, 2024 pm 05:38 PM
Breaking through the boundaries of traditional defect detection, 'Defect Spectrum' achieves ultra-high-precision and rich semantic industrial defect detection for the first time.
Jul 26, 2024 pm 05:38 PM
In modern manufacturing, accurate defect detection is not only the key to ensuring product quality, but also the core of improving production efficiency. However, existing defect detection datasets often lack the accuracy and semantic richness required for practical applications, resulting in models unable to identify specific defect categories or locations. In order to solve this problem, a top research team composed of Hong Kong University of Science and Technology Guangzhou and Simou Technology innovatively developed the "DefectSpectrum" data set, which provides detailed and semantically rich large-scale annotation of industrial defects. As shown in Table 1, compared with other industrial data sets, the "DefectSpectrum" data set provides the most defect annotations (5438 defect samples) and the most detailed defect classification (125 defect categories
 NVIDIA dialogue model ChatQA has evolved to version 2.0, with the context length mentioned at 128K
Jul 26, 2024 am 08:40 AM
NVIDIA dialogue model ChatQA has evolved to version 2.0, with the context length mentioned at 128K
Jul 26, 2024 am 08:40 AM
The open LLM community is an era when a hundred flowers bloom and compete. You can see Llama-3-70B-Instruct, QWen2-72B-Instruct, Nemotron-4-340B-Instruct, Mixtral-8x22BInstruct-v0.1 and many other excellent performers. Model. However, compared with proprietary large models represented by GPT-4-Turbo, open models still have significant gaps in many fields. In addition to general models, some open models that specialize in key areas have been developed, such as DeepSeek-Coder-V2 for programming and mathematics, and InternVL for visual-language tasks.
 Training with millions of crystal data to solve the crystallographic phase problem, the deep learning method PhAI is published in Science
Aug 08, 2024 pm 09:22 PM
Training with millions of crystal data to solve the crystallographic phase problem, the deep learning method PhAI is published in Science
Aug 08, 2024 pm 09:22 PM
Editor |KX To this day, the structural detail and precision determined by crystallography, from simple metals to large membrane proteins, are unmatched by any other method. However, the biggest challenge, the so-called phase problem, remains retrieving phase information from experimentally determined amplitudes. Researchers at the University of Copenhagen in Denmark have developed a deep learning method called PhAI to solve crystal phase problems. A deep learning neural network trained using millions of artificial crystal structures and their corresponding synthetic diffraction data can generate accurate electron density maps. The study shows that this deep learning-based ab initio structural solution method can solve the phase problem at a resolution of only 2 Angstroms, which is equivalent to only 10% to 20% of the data available at atomic resolution, while traditional ab initio Calculation
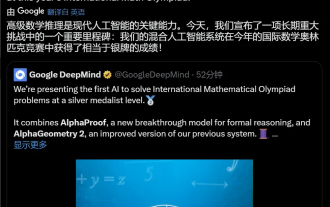 Google AI won the IMO Mathematical Olympiad silver medal, the mathematical reasoning model AlphaProof was launched, and reinforcement learning is so back
Jul 26, 2024 pm 02:40 PM
Google AI won the IMO Mathematical Olympiad silver medal, the mathematical reasoning model AlphaProof was launched, and reinforcement learning is so back
Jul 26, 2024 pm 02:40 PM
For AI, Mathematical Olympiad is no longer a problem. On Thursday, Google DeepMind's artificial intelligence completed a feat: using AI to solve the real question of this year's International Mathematical Olympiad IMO, and it was just one step away from winning the gold medal. The IMO competition that just ended last week had six questions involving algebra, combinatorics, geometry and number theory. The hybrid AI system proposed by Google got four questions right and scored 28 points, reaching the silver medal level. Earlier this month, UCLA tenured professor Terence Tao had just promoted the AI Mathematical Olympiad (AIMO Progress Award) with a million-dollar prize. Unexpectedly, the level of AI problem solving had improved to this level before July. Do the questions simultaneously on IMO. The most difficult thing to do correctly is IMO, which has the longest history, the largest scale, and the most negative
 Nature's point of view: The testing of artificial intelligence in medicine is in chaos. What should be done?
Aug 22, 2024 pm 04:37 PM
Nature's point of view: The testing of artificial intelligence in medicine is in chaos. What should be done?
Aug 22, 2024 pm 04:37 PM
Editor | ScienceAI Based on limited clinical data, hundreds of medical algorithms have been approved. Scientists are debating who should test the tools and how best to do so. Devin Singh witnessed a pediatric patient in the emergency room suffer cardiac arrest while waiting for treatment for a long time, which prompted him to explore the application of AI to shorten wait times. Using triage data from SickKids emergency rooms, Singh and colleagues built a series of AI models that provide potential diagnoses and recommend tests. One study showed that these models can speed up doctor visits by 22.3%, speeding up the processing of results by nearly 3 hours per patient requiring a medical test. However, the success of artificial intelligence algorithms in research only verifies this
 To provide a new scientific and complex question answering benchmark and evaluation system for large models, UNSW, Argonne, University of Chicago and other institutions jointly launched the SciQAG framework
Jul 25, 2024 am 06:42 AM
To provide a new scientific and complex question answering benchmark and evaluation system for large models, UNSW, Argonne, University of Chicago and other institutions jointly launched the SciQAG framework
Jul 25, 2024 am 06:42 AM
Editor |ScienceAI Question Answering (QA) data set plays a vital role in promoting natural language processing (NLP) research. High-quality QA data sets can not only be used to fine-tune models, but also effectively evaluate the capabilities of large language models (LLM), especially the ability to understand and reason about scientific knowledge. Although there are currently many scientific QA data sets covering medicine, chemistry, biology and other fields, these data sets still have some shortcomings. First, the data form is relatively simple, most of which are multiple-choice questions. They are easy to evaluate, but limit the model's answer selection range and cannot fully test the model's ability to answer scientific questions. In contrast, open-ended Q&A
 PRO | Why are large models based on MoE more worthy of attention?
Aug 07, 2024 pm 07:08 PM
PRO | Why are large models based on MoE more worthy of attention?
Aug 07, 2024 pm 07:08 PM
In 2023, almost every field of AI is evolving at an unprecedented speed. At the same time, AI is constantly pushing the technological boundaries of key tracks such as embodied intelligence and autonomous driving. Under the multi-modal trend, will the situation of Transformer as the mainstream architecture of AI large models be shaken? Why has exploring large models based on MoE (Mixed of Experts) architecture become a new trend in the industry? Can Large Vision Models (LVM) become a new breakthrough in general vision? ...From the 2023 PRO member newsletter of this site released in the past six months, we have selected 10 special interpretations that provide in-depth analysis of technological trends and industrial changes in the above fields to help you achieve your goals in the new year. be prepared. This interpretation comes from Week50 2023
 Automatically identify the best molecules and reduce synthesis costs. MIT develops a molecular design decision-making algorithm framework
Jun 22, 2024 am 06:43 AM
Automatically identify the best molecules and reduce synthesis costs. MIT develops a molecular design decision-making algorithm framework
Jun 22, 2024 am 06:43 AM
Editor | Ziluo AI’s use in streamlining drug discovery is exploding. Screen billions of candidate molecules for those that may have properties needed to develop new drugs. There are so many variables to consider, from material prices to the risk of error, that weighing the costs of synthesizing the best candidate molecules is no easy task, even if scientists use AI. Here, MIT researchers developed SPARROW, a quantitative decision-making algorithm framework, to automatically identify the best molecular candidates, thereby minimizing synthesis costs while maximizing the likelihood that the candidates have the desired properties. The algorithm also determined the materials and experimental steps needed to synthesize these molecules. SPARROW takes into account the cost of synthesizing a batch of molecules at once, since multiple candidate molecules are often available