 Technology peripherals
Technology peripherals
 AI
AI
 30 times higher than traditional methods, the Transformer deep learning model of the Chinese Academy of Sciences team predicts sugar-protein interaction sites
30 times higher than traditional methods, the Transformer deep learning model of the Chinese Academy of Sciences team predicts sugar-protein interaction sites
30 times higher than traditional methods, the Transformer deep learning model of the Chinese Academy of Sciences team predicts sugar-protein interaction sites

Saccharides are the most abundant organic substances in nature and are vital to life. Understanding how carbohydrates regulate proteins during physiological and pathological processes can provide opportunities to address key biological questions and develop new treatments.
However, the diversity and complexity of sugar molecules poses a challenge to experimentally identify sugar-protein binding and interaction sites.
Here, a team from the Chinese Academy of Sciences developed DeepGlycanSite, a deep learning model that is able to accurately predict sugar-binding sites on a given protein structure.
DeepGlycanSite integrates the geometric and evolutionary characteristics of proteins into a deep equivariant graph neural network with a Transformer architecture. Its performance significantly surpasses previous advanced methods and can effectively predict the binding sites of various sugar molecules.
Combined with mutagenesis studies, DeepGlycanSite reveals the guanosine-5'-bisphosphate recognition site of important G protein-coupled receptors.
These findings demonstrate the value of DeepGlycanSite for sugar binding site prediction and can provide insights into the molecular mechanisms behind sugar regulation of proteins of therapeutic importance.
The study was titled "Highly accurate carbohydrate-binding site prediction with DeepGlycanSite" and was published in "Nature Communications" on June 17, 2024.

Sugar is ubiquitous on the cell surface of all organisms. They interact with a variety of protein families such as lectins, antibodies, enzymes and transporters to regulate key biological processes such as immune response, cell differentiation and neural development. Understanding the interaction mechanism between carbohydrates and proteins is the basis for developing carbohydrate drugs.
However, the diversity and complexity of carbohydrate structures, especially the variability of their binding sites with proteins, pose challenges to the acquisition of experimental data and drug design.

Illustration: The complexity of sugar molecules and the diversity of sugar binding sites. (Source: paper)
In the past, traditional binding site prediction methods were not suitable for sugar molecules with complex structures and large changes in size. This, coupled with the scarcity of high-resolution structural data on sugar-protein complexes, limits the performance of predictive models.
In recent years, with the rapid development of Protein Data Bank (PDB) and open glycomics resources, the academic community has accumulated structural data of more than 19,000 such complexes. The increase in these high-quality data makes it possible to use AI technology to develop accurate sugar-binding site prediction models, which is expected to accelerate the discovery and optimization process of sugar drugs.
In the latest research, the Chinese Academy of Sciences team introduced DeepGlycanSite, a deep equivariant graph neural network (EGNN) model that can accurately predict sugar-binding sites with target protein structures.

Illustration: DeepGlycanSite overview. (Source: paper)
The team leveraged geometric features, such as directions and distances within and between residues, as well as evolutionary information, to present proteins as residue-level graphical representations in DeepGlycanSite. Combined with Transformer blocks with self-attention mechanism to enhance feature extraction and complex relationship discovery.
Researchers compared DeepGlycanSite to current state-of-the-art computational methods on an independent test set involving more than a hundred unique sugar-binding proteins.
The results show that the average Matthews correlation coefficient (MCC) of DeepGlycanSite (0.625) is more than 30 times that of StackCBPred (0.018), and far exceeds other sequence-based prediction methods.
Traditional ligand binding site methods may exclude binding sites for simple sugar molecules due to hydrophobicity or small size, while DeepGlycanSite can effectively identify these sites.

Illustration: Comparing model performance in predicting different sugar binding sites. (Source: paper)
Moreover, DeepGlycanSite also performs well in predicting multiple sugar-binding sites on proteins, which is of great value for understanding how multivalent glycoconjugates affect sugar-protein interactions and the regulation of biological processes. . For example, multivalent glycoconjugates are designed as chemical tools and drug candidates to influence the interaction between sugar molecules and lectins.
Different from traditional methods that only use protein sequence or structural information, DeepGlycanSite fully considers the geometric information and evolutionary characteristics of the protein, which may be the key factor for its excellent performance.
Additionally, given the chemical structure of a query sugar molecule, DeepGlycanSite can predict its specific binding site.

Illustration: Query the prediction of specific binding sites of sugars. (Source: Paper)
Researchers explored the application of DeepGlycanSite to functionally important G protein-coupled receptors (GPCRs). Using the protein structure and carbohydrate chemical structure predicted by AlphaFold2, DeepGlycanSite successfully detected the specific binding site of GDP-Fuc on human P2Y14.

While the quality of AlphaFold2’s predicted side chains needs to be improved, DeepGlycanSite relies less on protein structure accuracy and is able to use predicted protein structures to provide insights into sugar-protein interactions.
In summary, the validation of DeepGlycanSite in independent test sets and in vitro case studies shows that it is an effective tool for sugar binding site prediction. Researchers can use DeepGlycanSite to predict sugar-binding pockets on target proteins, thereby advancing understanding of sugar-protein interactions.
Saccharides play a key role in biological functions. DeepGlycanSite not only helps analyze the biological functions of sugar molecules and sugar-binding proteins, but also provides a powerful tool for the development of sugar drugs.
Paper link:https://www.nature.com/articles/s41467-024-49516-2
The above is the detailed content of 30 times higher than traditional methods, the Transformer deep learning model of the Chinese Academy of Sciences team predicts sugar-protein interaction sites. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

AI Hentai Generator
Generate AI Hentai for free.

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1376
1376
 52
52
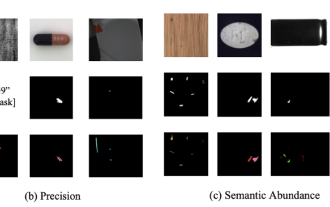 Breaking through the boundaries of traditional defect detection, 'Defect Spectrum' achieves ultra-high-precision and rich semantic industrial defect detection for the first time.
Jul 26, 2024 pm 05:38 PM
Breaking through the boundaries of traditional defect detection, 'Defect Spectrum' achieves ultra-high-precision and rich semantic industrial defect detection for the first time.
Jul 26, 2024 pm 05:38 PM
In modern manufacturing, accurate defect detection is not only the key to ensuring product quality, but also the core of improving production efficiency. However, existing defect detection datasets often lack the accuracy and semantic richness required for practical applications, resulting in models unable to identify specific defect categories or locations. In order to solve this problem, a top research team composed of Hong Kong University of Science and Technology Guangzhou and Simou Technology innovatively developed the "DefectSpectrum" data set, which provides detailed and semantically rich large-scale annotation of industrial defects. As shown in Table 1, compared with other industrial data sets, the "DefectSpectrum" data set provides the most defect annotations (5438 defect samples) and the most detailed defect classification (125 defect categories
 NVIDIA dialogue model ChatQA has evolved to version 2.0, with the context length mentioned at 128K
Jul 26, 2024 am 08:40 AM
NVIDIA dialogue model ChatQA has evolved to version 2.0, with the context length mentioned at 128K
Jul 26, 2024 am 08:40 AM
The open LLM community is an era when a hundred flowers bloom and compete. You can see Llama-3-70B-Instruct, QWen2-72B-Instruct, Nemotron-4-340B-Instruct, Mixtral-8x22BInstruct-v0.1 and many other excellent performers. Model. However, compared with proprietary large models represented by GPT-4-Turbo, open models still have significant gaps in many fields. In addition to general models, some open models that specialize in key areas have been developed, such as DeepSeek-Coder-V2 for programming and mathematics, and InternVL for visual-language tasks.
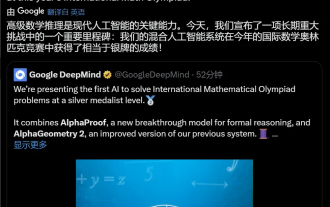 Google AI won the IMO Mathematical Olympiad silver medal, the mathematical reasoning model AlphaProof was launched, and reinforcement learning is so back
Jul 26, 2024 pm 02:40 PM
Google AI won the IMO Mathematical Olympiad silver medal, the mathematical reasoning model AlphaProof was launched, and reinforcement learning is so back
Jul 26, 2024 pm 02:40 PM
For AI, Mathematical Olympiad is no longer a problem. On Thursday, Google DeepMind's artificial intelligence completed a feat: using AI to solve the real question of this year's International Mathematical Olympiad IMO, and it was just one step away from winning the gold medal. The IMO competition that just ended last week had six questions involving algebra, combinatorics, geometry and number theory. The hybrid AI system proposed by Google got four questions right and scored 28 points, reaching the silver medal level. Earlier this month, UCLA tenured professor Terence Tao had just promoted the AI Mathematical Olympiad (AIMO Progress Award) with a million-dollar prize. Unexpectedly, the level of AI problem solving had improved to this level before July. Do the questions simultaneously on IMO. The most difficult thing to do correctly is IMO, which has the longest history, the largest scale, and the most negative
 Nature's point of view: The testing of artificial intelligence in medicine is in chaos. What should be done?
Aug 22, 2024 pm 04:37 PM
Nature's point of view: The testing of artificial intelligence in medicine is in chaos. What should be done?
Aug 22, 2024 pm 04:37 PM
Editor | ScienceAI Based on limited clinical data, hundreds of medical algorithms have been approved. Scientists are debating who should test the tools and how best to do so. Devin Singh witnessed a pediatric patient in the emergency room suffer cardiac arrest while waiting for treatment for a long time, which prompted him to explore the application of AI to shorten wait times. Using triage data from SickKids emergency rooms, Singh and colleagues built a series of AI models that provide potential diagnoses and recommend tests. One study showed that these models can speed up doctor visits by 22.3%, speeding up the processing of results by nearly 3 hours per patient requiring a medical test. However, the success of artificial intelligence algorithms in research only verifies this
 Training with millions of crystal data to solve the crystallographic phase problem, the deep learning method PhAI is published in Science
Aug 08, 2024 pm 09:22 PM
Training with millions of crystal data to solve the crystallographic phase problem, the deep learning method PhAI is published in Science
Aug 08, 2024 pm 09:22 PM
Editor |KX To this day, the structural detail and precision determined by crystallography, from simple metals to large membrane proteins, are unmatched by any other method. However, the biggest challenge, the so-called phase problem, remains retrieving phase information from experimentally determined amplitudes. Researchers at the University of Copenhagen in Denmark have developed a deep learning method called PhAI to solve crystal phase problems. A deep learning neural network trained using millions of artificial crystal structures and their corresponding synthetic diffraction data can generate accurate electron density maps. The study shows that this deep learning-based ab initio structural solution method can solve the phase problem at a resolution of only 2 Angstroms, which is equivalent to only 10% to 20% of the data available at atomic resolution, while traditional ab initio Calculation
 To provide a new scientific and complex question answering benchmark and evaluation system for large models, UNSW, Argonne, University of Chicago and other institutions jointly launched the SciQAG framework
Jul 25, 2024 am 06:42 AM
To provide a new scientific and complex question answering benchmark and evaluation system for large models, UNSW, Argonne, University of Chicago and other institutions jointly launched the SciQAG framework
Jul 25, 2024 am 06:42 AM
Editor |ScienceAI Question Answering (QA) data set plays a vital role in promoting natural language processing (NLP) research. High-quality QA data sets can not only be used to fine-tune models, but also effectively evaluate the capabilities of large language models (LLM), especially the ability to understand and reason about scientific knowledge. Although there are currently many scientific QA data sets covering medicine, chemistry, biology and other fields, these data sets still have some shortcomings. First, the data form is relatively simple, most of which are multiple-choice questions. They are easy to evaluate, but limit the model's answer selection range and cannot fully test the model's ability to answer scientific questions. In contrast, open-ended Q&A
 Automatically identify the best molecules and reduce synthesis costs. MIT develops a molecular design decision-making algorithm framework
Jun 22, 2024 am 06:43 AM
Automatically identify the best molecules and reduce synthesis costs. MIT develops a molecular design decision-making algorithm framework
Jun 22, 2024 am 06:43 AM
Editor | Ziluo AI’s use in streamlining drug discovery is exploding. Screen billions of candidate molecules for those that may have properties needed to develop new drugs. There are so many variables to consider, from material prices to the risk of error, that weighing the costs of synthesizing the best candidate molecules is no easy task, even if scientists use AI. Here, MIT researchers developed SPARROW, a quantitative decision-making algorithm framework, to automatically identify the best molecular candidates, thereby minimizing synthesis costs while maximizing the likelihood that the candidates have the desired properties. The algorithm also determined the materials and experimental steps needed to synthesize these molecules. SPARROW takes into account the cost of synthesizing a batch of molecules at once, since multiple candidate molecules are often available
 SOTA performance, Xiamen multi-modal protein-ligand affinity prediction AI method, combines molecular surface information for the first time
Jul 17, 2024 pm 06:37 PM
SOTA performance, Xiamen multi-modal protein-ligand affinity prediction AI method, combines molecular surface information for the first time
Jul 17, 2024 pm 06:37 PM
Editor | KX In the field of drug research and development, accurately and effectively predicting the binding affinity of proteins and ligands is crucial for drug screening and optimization. However, current studies do not take into account the important role of molecular surface information in protein-ligand interactions. Based on this, researchers from Xiamen University proposed a novel multi-modal feature extraction (MFE) framework, which for the first time combines information on protein surface, 3D structure and sequence, and uses a cross-attention mechanism to compare different modalities. feature alignment. Experimental results demonstrate that this method achieves state-of-the-art performance in predicting protein-ligand binding affinities. Furthermore, ablation studies demonstrate the effectiveness and necessity of protein surface information and multimodal feature alignment within this framework. Related research begins with "S



