

Large language models (LLMs) such as OpenAI’s GPT and Meta AI’s Llama are increasingly recognized for their potential in the field of chemoinformatics, especially in understanding simplified molecular input line input systems ( SMILES) aspect. These LLMs are also able to decode SMILES strings into vector representations.
Researchers at the University of Windsor in Canada compared the performance of pre-trained models on GPT and Llama with SMILES for embedding SMILES strings in downstream tasks, focusing on two key applications: molecular property prediction and drug-drug Interaction prediction.
The study was titled "Can large language models understand molecules?" and was published in "BMC Bioinformatics" on June 25, 2024.
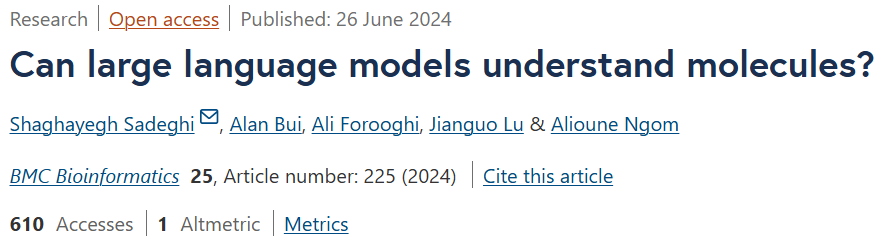
Molecular embedding is a crucial task in drug discovery and is widely used in molecular property prediction, drug-target interaction (DTI) prediction and drug-drug interaction Function (DDI) prediction and other related tasks.
2. Molecular embedding technology
Molecular embedding technology can learn features from molecular graphs encoding molecular structural connection information or line annotations of their structures, such as the popular SMILES representation.
3. Molecular embeddings in SMILES strings
Molecular embeddings via SMILES strings have evolved in tandem with advances in language modeling, from static word embeddings to contextualized pre-trained models. These embedding techniques aim to capture relevant structural and chemical information in a compact numerical representation.
The basic assumption is that molecules with similar structures behave in similar ways. This enables machine learning algorithms to process and analyze molecular structures for property prediction and drug discovery tasks.
With breakthroughs in LLM, a prominent question is whether LLM can understand molecules and make inferences based on molecular data?
More specifically, can LLM produce high-quality semantic representations?
Shaghayegh Sadeghi, Alioune Ngom Jianguo Lu and others at the University of Windsor further explored the ability of these models to effectively embed SMILES. Currently, this capability is underexplored, perhaps in part because of the cost of the API calls.
Researchers found that SMILES embeddings generated using Llama performed better than SMILES embeddings generated using GPT in both molecular property and DDI prediction tasks.
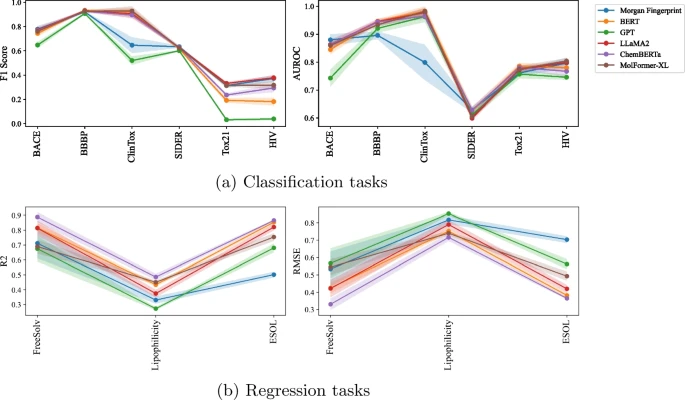

The above is the detailed content of Llama molecule embedding is better than GPT, can LLM understand molecules? Meta defeated OpenAI in this round. For more information, please follow other related articles on the PHP Chinese website!




