Technology peripherals
Technology peripherals
 AI
AI
 Generating 394,760 protein representations, Harvard team develops AI model to fully understand protein context
Generating 394,760 protein representations, Harvard team develops AI model to fully understand protein context
Generating 394,760 protein representations, Harvard team develops AI model to fully understand protein context

Understanding protein function and developing molecular therapies requires identifying the cell types in which proteins play a role and analyzing the interactions between proteins.
However, modeling protein interactions across biological contexts remains challenging for existing algorithms.
In the latest study, researchers at Harvard Medical School developed PINNACLE, a geometric deep learning method for generating context-aware protein representations.
PINNACLE leverages multi-organ single cell atlases to learn on contextualized protein interaction networks, generating 394,760 protein representations from 156 cell type contexts across 24 tissues.
The study was titled "Contextual AI models for single-cell protein biology" and was published in "Nature Methods" on July 22, 2024.

- Proteins are the basic functional units of cells and achieve biological functions through interactions.
- High-throughput technologies have driven the mapping of protein interaction networks and improved understanding of protein structure, function and target design through computational methods.
- Indicates that the learning method integrates molecular cell atlases, can analyze protein interaction networks, and expands the understanding of protein functions.
Context-dependent protein function
- Proteins play different roles in different biological contexts, and gene expression and function vary according to health and disease states.
- Background-free protein means that functional changes between cell types cannot be identified, affecting prediction accuracy.
Single Cell Gene Expression and Protein Network
- Sequencing technology measures single cell gene expression, paving the way for solving context-dependent problems.
- Attention-based deep learning can focus on large inputs and learn important elements in the context.
- Single cell atlas can enhance the mapping of gene regulatory networks related to disease progression and reveal targets.
PINNACLE MODEL
- There are still challenges in integrating protein-coding gene expression into protein interaction networks.
- PINNACLE model provides context-specific understanding of proteins.
- PINNACLE is a geometric deep learning model that generates protein representations by analyzing protein interactions in cellular environments.
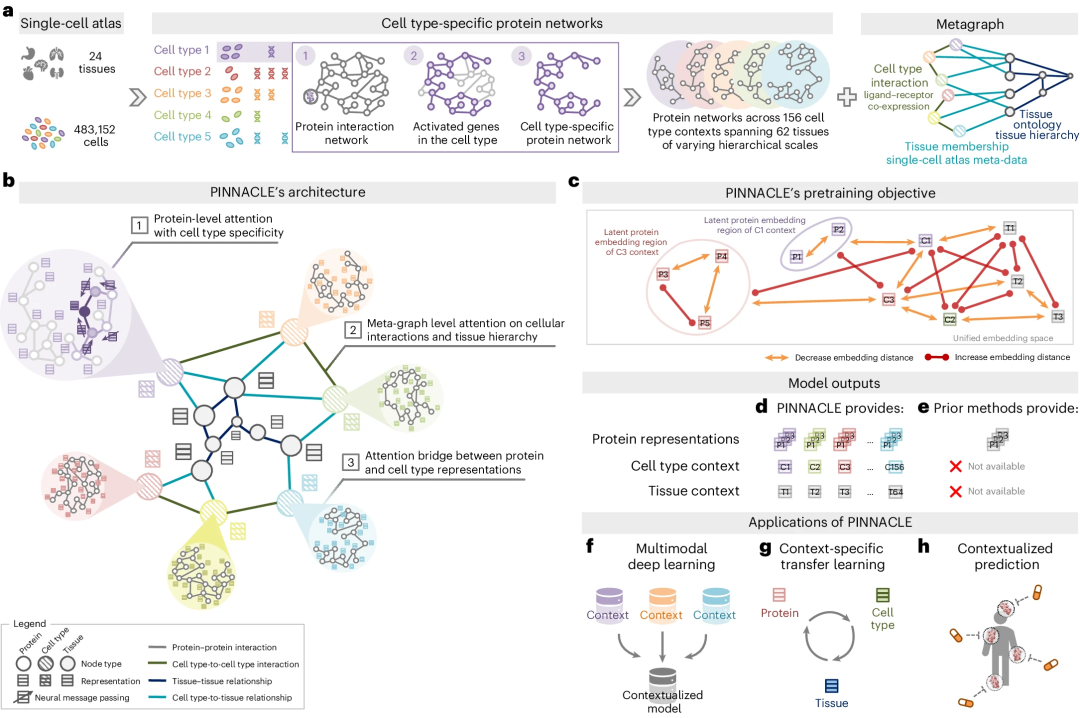
1. PINNACLE Overview
Illustration: PINNACLE Overview. (Source: Paper)
2. Contextualized Protein Representation
PINNACLE is trained on an integrated context-aware PPI network, supplemented by a network that captures cellular interactions and tissue hierarchies, to generate protein representations customized for cell types .
3. Multi-scale representation
Unlike context-free models, PINNACLE generates multiple representations for each protein, depending on its cell type context. Additionally, PINNACLE generates cell type context and tissue-level representations.
4. Multi-scale learning
PINNACLE learns the topology of proteins, cell types and tissues by optimizing a unified latent representation space.
5. Context-Aware Models
PINNACLE integrates context-specific data into a single model and transfers knowledge between protein, cell type and tissue-level data.
6. Embedding Space
To inject cellular and tissue information into the embedding space, PINNACLE employs protein, cell type, and tissue level attention.
7. Physical interaction mapping
Physically interacting protein pairs are tightly embedded in the embedding space.
8. Cell Type Environment
Proteins are embedded near their cell type environment.
9. Graph Neural Network Propagation
PINNACLE propagates information between proteins, cell types and tissues using an attention mechanism customized for each node and edge type.

PINNACLE’s cell-type and tissue-specific pre-training tasks rely entirely on self-supervised link prediction to facilitate learning of cell and tissue organization. The topology of cell types and tissues is passed to the protein representation through an attention bridging mechanism, effectively reinforcing tissue and cell organization onto the protein representation.
PINNACLE’s contextualized protein representation captures the structure of context-aware protein interaction networks. The regional arrangement of these contextualized protein representations in latent space reflects the cellular and tissue organization represented by the metagraph. This will lead to a comprehensive and context-specific representation of proteins within a unified cell-type and tissue-specific framework.
With 394,760 contextualized protein representations generated by PINNACLE, each of which is cell type specific, researchers demonstrate PINNACLE's ability to combine protein interactions with the underlying protein-coding gene transcriptomes of 156 cell type contexts.
PINNACLE’s embedding space reflects cellular and tissue structures, enabling zero-shot retrieval of tissue hierarchies. Pretrained protein representations can be adapted to downstream tasks: enhance 3D structure-based representations to resolve immuno-oncology protein interactions and study the effects of drugs on different cell types.
PINNACLE outperforms state-of-the-art models in specifying therapeutic targets for rheumatoid arthritis and inflammatory bowel disease, and has higher predictive power than context-free models to pinpoint cell type context. PINNACLE's ability to adapt its output to the environment in which it operates paves the way for large-scale context-specific predictions in biology.
Paper link: https://www.nature.com/articles/s41592-024-02341-3
The above is the detailed content of Generating 394,760 protein representations, Harvard team develops AI model to fully understand protein context. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1666
1666
 14
14
 1425
1425
 52
52
 1328
1328
 25
25
 1273
1273
 29
29
 1253
1253
 24
24
 Breaking through the boundaries of traditional defect detection, 'Defect Spectrum' achieves ultra-high-precision and rich semantic industrial defect detection for the first time.
Jul 26, 2024 pm 05:38 PM
Breaking through the boundaries of traditional defect detection, 'Defect Spectrum' achieves ultra-high-precision and rich semantic industrial defect detection for the first time.
Jul 26, 2024 pm 05:38 PM
In modern manufacturing, accurate defect detection is not only the key to ensuring product quality, but also the core of improving production efficiency. However, existing defect detection datasets often lack the accuracy and semantic richness required for practical applications, resulting in models unable to identify specific defect categories or locations. In order to solve this problem, a top research team composed of Hong Kong University of Science and Technology Guangzhou and Simou Technology innovatively developed the "DefectSpectrum" data set, which provides detailed and semantically rich large-scale annotation of industrial defects. As shown in Table 1, compared with other industrial data sets, the "DefectSpectrum" data set provides the most defect annotations (5438 defect samples) and the most detailed defect classification (125 defect categories
 Beyond ORB-SLAM3! SL-SLAM: Low light, severe jitter and weak texture scenes are all handled
May 30, 2024 am 09:35 AM
Beyond ORB-SLAM3! SL-SLAM: Low light, severe jitter and weak texture scenes are all handled
May 30, 2024 am 09:35 AM
Written previously, today we discuss how deep learning technology can improve the performance of vision-based SLAM (simultaneous localization and mapping) in complex environments. By combining deep feature extraction and depth matching methods, here we introduce a versatile hybrid visual SLAM system designed to improve adaptation in challenging scenarios such as low-light conditions, dynamic lighting, weakly textured areas, and severe jitter. sex. Our system supports multiple modes, including extended monocular, stereo, monocular-inertial, and stereo-inertial configurations. In addition, it also analyzes how to combine visual SLAM with deep learning methods to inspire other research. Through extensive experiments on public datasets and self-sampled data, we demonstrate the superiority of SL-SLAM in terms of positioning accuracy and tracking robustness.
 Training with millions of crystal data to solve the crystallographic phase problem, the deep learning method PhAI is published in Science
Aug 08, 2024 pm 09:22 PM
Training with millions of crystal data to solve the crystallographic phase problem, the deep learning method PhAI is published in Science
Aug 08, 2024 pm 09:22 PM
Editor |KX To this day, the structural detail and precision determined by crystallography, from simple metals to large membrane proteins, are unmatched by any other method. However, the biggest challenge, the so-called phase problem, remains retrieving phase information from experimentally determined amplitudes. Researchers at the University of Copenhagen in Denmark have developed a deep learning method called PhAI to solve crystal phase problems. A deep learning neural network trained using millions of artificial crystal structures and their corresponding synthetic diffraction data can generate accurate electron density maps. The study shows that this deep learning-based ab initio structural solution method can solve the phase problem at a resolution of only 2 Angstroms, which is equivalent to only 10% to 20% of the data available at atomic resolution, while traditional ab initio Calculation
 NVIDIA dialogue model ChatQA has evolved to version 2.0, with the context length mentioned at 128K
Jul 26, 2024 am 08:40 AM
NVIDIA dialogue model ChatQA has evolved to version 2.0, with the context length mentioned at 128K
Jul 26, 2024 am 08:40 AM
The open LLM community is an era when a hundred flowers bloom and compete. You can see Llama-3-70B-Instruct, QWen2-72B-Instruct, Nemotron-4-340B-Instruct, Mixtral-8x22BInstruct-v0.1 and many other excellent performers. Model. However, compared with proprietary large models represented by GPT-4-Turbo, open models still have significant gaps in many fields. In addition to general models, some open models that specialize in key areas have been developed, such as DeepSeek-Coder-V2 for programming and mathematics, and InternVL for visual-language tasks.
 Google AI won the IMO Mathematical Olympiad silver medal, the mathematical reasoning model AlphaProof was launched, and reinforcement learning is so back
Jul 26, 2024 pm 02:40 PM
Google AI won the IMO Mathematical Olympiad silver medal, the mathematical reasoning model AlphaProof was launched, and reinforcement learning is so back
Jul 26, 2024 pm 02:40 PM
For AI, Mathematical Olympiad is no longer a problem. On Thursday, Google DeepMind's artificial intelligence completed a feat: using AI to solve the real question of this year's International Mathematical Olympiad IMO, and it was just one step away from winning the gold medal. The IMO competition that just ended last week had six questions involving algebra, combinatorics, geometry and number theory. The hybrid AI system proposed by Google got four questions right and scored 28 points, reaching the silver medal level. Earlier this month, UCLA tenured professor Terence Tao had just promoted the AI Mathematical Olympiad (AIMO Progress Award) with a million-dollar prize. Unexpectedly, the level of AI problem solving had improved to this level before July. Do the questions simultaneously on IMO. The most difficult thing to do correctly is IMO, which has the longest history, the largest scale, and the most negative
 PRO | Why are large models based on MoE more worthy of attention?
Aug 07, 2024 pm 07:08 PM
PRO | Why are large models based on MoE more worthy of attention?
Aug 07, 2024 pm 07:08 PM
In 2023, almost every field of AI is evolving at an unprecedented speed. At the same time, AI is constantly pushing the technological boundaries of key tracks such as embodied intelligence and autonomous driving. Under the multi-modal trend, will the situation of Transformer as the mainstream architecture of AI large models be shaken? Why has exploring large models based on MoE (Mixed of Experts) architecture become a new trend in the industry? Can Large Vision Models (LVM) become a new breakthrough in general vision? ...From the 2023 PRO member newsletter of this site released in the past six months, we have selected 10 special interpretations that provide in-depth analysis of technological trends and industrial changes in the above fields to help you achieve your goals in the new year. be prepared. This interpretation comes from Week50 2023
 AlphaFold 3 is launched, comprehensively predicting the interactions and structures of proteins and all living molecules, with far greater accuracy than ever before
Jul 16, 2024 am 12:08 AM
AlphaFold 3 is launched, comprehensively predicting the interactions and structures of proteins and all living molecules, with far greater accuracy than ever before
Jul 16, 2024 am 12:08 AM
Editor | Radish Skin Since the release of the powerful AlphaFold2 in 2021, scientists have been using protein structure prediction models to map various protein structures within cells, discover drugs, and draw a "cosmic map" of every known protein interaction. . Just now, Google DeepMind released the AlphaFold3 model, which can perform joint structure predictions for complexes including proteins, nucleic acids, small molecules, ions and modified residues. The accuracy of AlphaFold3 has been significantly improved compared to many dedicated tools in the past (protein-ligand interaction, protein-nucleic acid interaction, antibody-antigen prediction). This shows that within a single unified deep learning framework, it is possible to achieve
 To provide a new scientific and complex question answering benchmark and evaluation system for large models, UNSW, Argonne, University of Chicago and other institutions jointly launched the SciQAG framework
Jul 25, 2024 am 06:42 AM
To provide a new scientific and complex question answering benchmark and evaluation system for large models, UNSW, Argonne, University of Chicago and other institutions jointly launched the SciQAG framework
Jul 25, 2024 am 06:42 AM
Editor |ScienceAI Question Answering (QA) data set plays a vital role in promoting natural language processing (NLP) research. High-quality QA data sets can not only be used to fine-tune models, but also effectively evaluate the capabilities of large language models (LLM), especially the ability to understand and reason about scientific knowledge. Although there are currently many scientific QA data sets covering medicine, chemistry, biology and other fields, these data sets still have some shortcomings. First, the data form is relatively simple, most of which are multiple-choice questions. They are easy to evaluate, but limit the model's answer selection range and cannot fully test the model's ability to answer scientific questions. In contrast, open-ended Q&A