 Technology peripherals
Technology peripherals
 AI
AI
 AI designs protein 'switches' from scratch, an amazing breakthrough in protein design, David Baker's research is published in Nature
AI designs protein 'switches' from scratch, an amazing breakthrough in protein design, David Baker's research is published in Nature
AI designs protein 'switches' from scratch, an amazing breakthrough in protein design, David Baker's research is published in Nature

In life, it’s easy to turn on a light or adjust the light. But systems that achieve similar control of biomolecular functions are complex and poorly understood.
In biology, protein functions are turned on and off in complex ways. Allosteric regulation is one of the important biological regulatory mechanisms and is crucial for healthy metabolism and cell signaling. But creating allostery in synthetic protein systems has always presented significant challenges.
Recently, David Baker’s team at the University of Washington designed a protein that can switch between assembly and disassembly reliably and accurately through allosteric control. Using AI to design new proteins that do not exist in nature, researchers have engineered multiple dynamic protein arrangements.
David Baker said: "By designing proteins that can be assembled and disassembled on command, we are paving the way for future biotechnologies that may rival the complexity of nature."
# 🎜🎜#Arvind Pillai, first author and corresponding author of the paper, said: "One of the key innovations of this study is the design of protein assemblies that can switch between different oligomer states, such as dimers. , rings and cages in response to effector molecules, this ability to remotely control protein structure opens up the possibility of developing adaptive biomaterials and drug delivery systems. A stunning breakthrough in design." The relevant research was titled "De novo design of allosterically switchable protein assemblies" and was published in "Nature" on August 14.Allostery and de novo design
 Paper link: https://www.nature.com/articles/s41586-024-07813-2#🎜 🎜#
Paper link: https://www.nature.com/articles/s41586-024-07813-2#🎜 🎜#
- Proteins designed to change structure and function in response to specific molecular signals are called
- Allosteric Regulation#🎜🎜 #.
De novo design
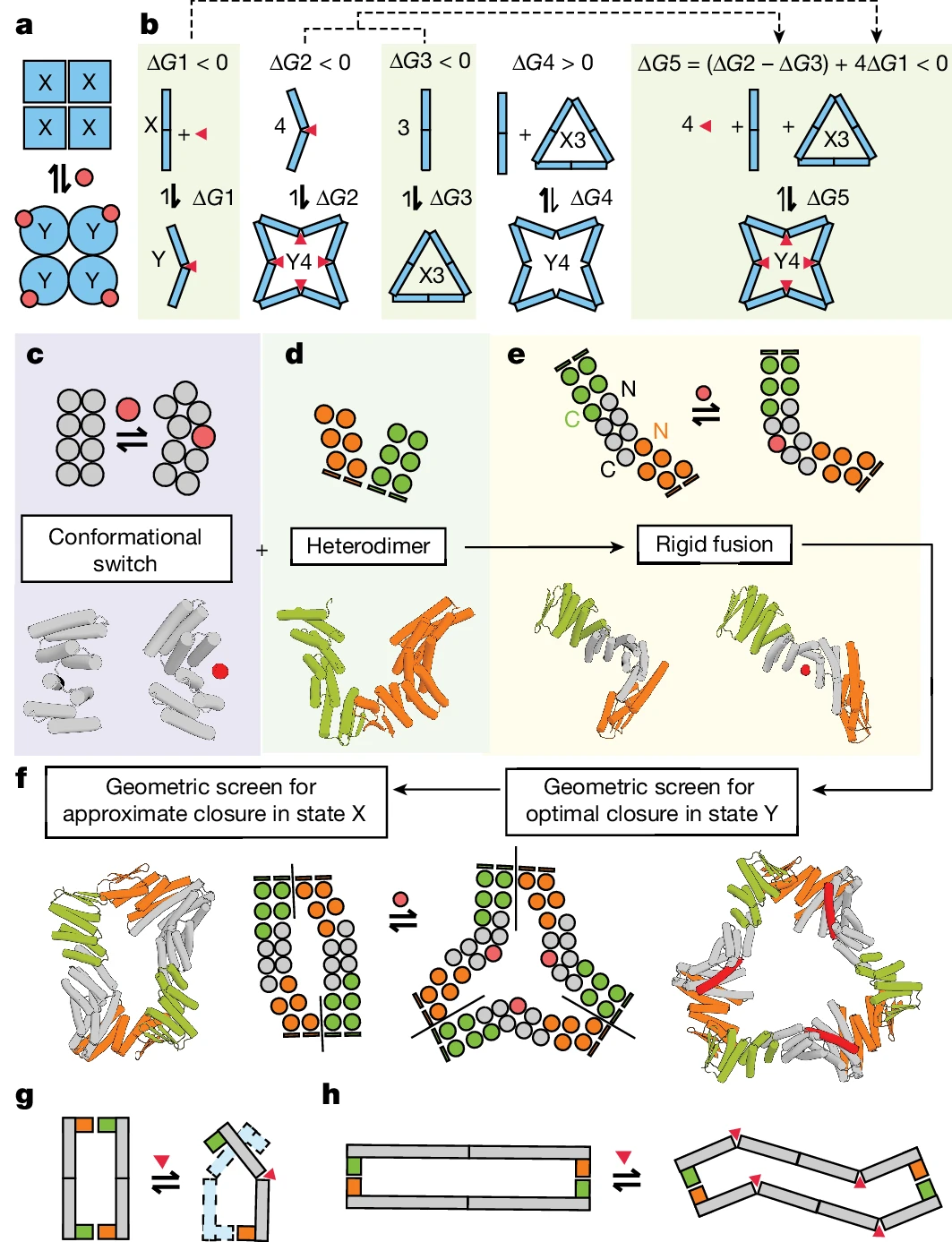
De novo designed proteins expand the repertoire of naturally evolved properties, opening the door to more controllable control of protein function.- Inspiration: Monod-Wyman-Changeux (MWC) collaborative model
- allosteric
- .
Design strategy
- Use the MWC model as a starting point for design
- Utilize protein structure prediction tools# 🎜🎜#
- Structural Switch
- Sewing protein modules in a structurally and energetically feasible way
- #🎜 🎜#Illustration: Design strategies for building switchable oligomers. (Source: paper)
-
Researchers demonstrated the application of RFdiffusion, ProteinMPNN and other design tools, For creating a series of dynamic and conformational switching protein assemblies. By combining two-state hinges and customized protein-protein interaction modules, the resulting assemblies are significantly different from any previously seen, expanding the possibilities of synthetic biology. Research results:
Research results:
Key Innovation:
A key innovation in this research is the design of protein assemblies. In addition to structural versatility, the team also achieved high-affinity binding between the new protein and its effectors, ensuring reliable programmed allosteric control. "For this project, we used specific peptides as effectors, but any type of molecule can be used in Protein allostery occurs under the right conditions," added co-author Abbas Idris, a graduate student at the University of Washington.Illustration: Design of allosterically controlled ring assembly. (Source: paper) The researchers synthesized a number of designed proteins and then characterized the protein structure and the switching behavior caused by the binding of effector molecules. Nearly 40% of synthetic proteins designed to switch between ring assemblies composed of varying numbers of protomers are water-soluble and display the expected protomer stoichiometry.
Furthermore, the number of effectors bound to a protein follows the MWC model: all binding sites are filled, or none at all. In other words, homologous effector binding is highly cooperative and the resulting assembly does not contain a mixture of R and T protomers.
Illustration: A protein switches between assembly states by design. (Source: Nature)
Going one step further, the researchers designed proteins containing double hinges (two hinges connected by short loops) with the goal of creating structures that respond to effector binding without changing the number of protomers in the protein assembly. And the protein changes its 3D structure. Sure enough, these proteins functioned as expected, reproducing the dominant behavior of naturally occurring allosteric proteins such as hemoglobin. Finally, the researchers also designed protomers that assemble or disassemble when bound to effector molecules.
Specific de novo protein assemblies designed in the study include rings formed by the dimerization of two monomers, which upon assembly trigger light output for biosensing applications, and cage-like structures, which undergo controlled disassembly for Release the payload for drug delivery. These protein dynamics were experimentally verified in vitro by size exclusion chromatography, mass spectrometry, and electron microscopy.
Pillai emphasized that ring structures exhibit additional precise properties such as cooperativity, a phenomenon exhibited by natural systems (e.g. blood proteins, hemoglobin). In synergistic systems, the binding of one molecule enhances the binding of other molecules, creating rapid on-off reactions that are critical for precise control, such as capturing oxygen in the lungs and releasing it into tissues.
"Historically, in the laboratory, we've done a lot to control the affinity of binding a substance, such as binding it tighter and tighter. But that's not the only aspect relevant to biological systems," Pillai said. "Sometimes you want to be able to bind over a very narrow concentration range." To validate the design, the researchers characterized more than 20 protein assemblies using negative staining and cryo-electron microscopy. "This allowed us to confirm which designs formed as expected and observe how these assemblies changed their structure when effector molecules were introduced," explains Dr. Andrew Borst, head of the IPD Electron Microscopy Research Core.
Illustration: Structure characterization of sr312 and sr322 by electron microscopy. (Source: paper) 
Designed components include nanoscale containers that can be opened and closed remotely. Such systems could lead to novel drug delivery vehicles with advanced control mechanisms, including devices that sequester cell-killing drugs until they encounter tumors.
This research paves the way for the design of allosterically controlled functions beyond protein assembly and disassembly, such as regulating enzyme activity for metabolic functions and nanomachines that can convert energy into mechanical work, similar to the proteins responsible for cell movement. Actin and myosin.
“The next step is to determine whether we can form interactions with small molecules and accurately catalyze reactions, which is a more challenging frontier for the entire field,” Pillai said.
Going forward, the research team seeks to evaluate these engineered protein dynamics in a broader biological context. Future work includes installing these engineered features on cell surfaces in tissue culture, providing valuable tools for feedback control in therapeutics such as adoptive cell therapy.
Reference content:
https://www.bakerlab.org/2024/08/14/morphing-protein-assemblies-by-design/- https://www.genengnews.com/topics/artificial- intelligence/ai-designed-proteins-morph-on-demand-for-steerable-functionality/
- https://www.nature.com/articles/d41586-024-02242-7
The above is the detailed content of AI designs protein 'switches' from scratch, an amazing breakthrough in protein design, David Baker's research is published in Nature. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1664
1664
 14
14
 1422
1422
 52
52
 1316
1316
 25
25
 1267
1267
 29
29
 1239
1239
 24
24
 How to understand DMA operations in C?
Apr 28, 2025 pm 10:09 PM
How to understand DMA operations in C?
Apr 28, 2025 pm 10:09 PM
DMA in C refers to DirectMemoryAccess, a direct memory access technology, allowing hardware devices to directly transmit data to memory without CPU intervention. 1) DMA operation is highly dependent on hardware devices and drivers, and the implementation method varies from system to system. 2) Direct access to memory may bring security risks, and the correctness and security of the code must be ensured. 3) DMA can improve performance, but improper use may lead to degradation of system performance. Through practice and learning, we can master the skills of using DMA and maximize its effectiveness in scenarios such as high-speed data transmission and real-time signal processing.
 How to use the chrono library in C?
Apr 28, 2025 pm 10:18 PM
How to use the chrono library in C?
Apr 28, 2025 pm 10:18 PM
Using the chrono library in C can allow you to control time and time intervals more accurately. Let's explore the charm of this library. C's chrono library is part of the standard library, which provides a modern way to deal with time and time intervals. For programmers who have suffered from time.h and ctime, chrono is undoubtedly a boon. It not only improves the readability and maintainability of the code, but also provides higher accuracy and flexibility. Let's start with the basics. The chrono library mainly includes the following key components: std::chrono::system_clock: represents the system clock, used to obtain the current time. std::chron
 Quantitative Exchange Ranking 2025 Top 10 Recommendations for Digital Currency Quantitative Trading APPs
Apr 30, 2025 pm 07:24 PM
Quantitative Exchange Ranking 2025 Top 10 Recommendations for Digital Currency Quantitative Trading APPs
Apr 30, 2025 pm 07:24 PM
The built-in quantization tools on the exchange include: 1. Binance: Provides Binance Futures quantitative module, low handling fees, and supports AI-assisted transactions. 2. OKX (Ouyi): Supports multi-account management and intelligent order routing, and provides institutional-level risk control. The independent quantitative strategy platforms include: 3. 3Commas: drag-and-drop strategy generator, suitable for multi-platform hedging arbitrage. 4. Quadency: Professional-level algorithm strategy library, supporting customized risk thresholds. 5. Pionex: Built-in 16 preset strategy, low transaction fee. Vertical domain tools include: 6. Cryptohopper: cloud-based quantitative platform, supporting 150 technical indicators. 7. Bitsgap:
 How to handle high DPI display in C?
Apr 28, 2025 pm 09:57 PM
How to handle high DPI display in C?
Apr 28, 2025 pm 09:57 PM
Handling high DPI display in C can be achieved through the following steps: 1) Understand DPI and scaling, use the operating system API to obtain DPI information and adjust the graphics output; 2) Handle cross-platform compatibility, use cross-platform graphics libraries such as SDL or Qt; 3) Perform performance optimization, improve performance through cache, hardware acceleration, and dynamic adjustment of the details level; 4) Solve common problems, such as blurred text and interface elements are too small, and solve by correctly applying DPI scaling.
 What is real-time operating system programming in C?
Apr 28, 2025 pm 10:15 PM
What is real-time operating system programming in C?
Apr 28, 2025 pm 10:15 PM
C performs well in real-time operating system (RTOS) programming, providing efficient execution efficiency and precise time management. 1) C Meet the needs of RTOS through direct operation of hardware resources and efficient memory management. 2) Using object-oriented features, C can design a flexible task scheduling system. 3) C supports efficient interrupt processing, but dynamic memory allocation and exception processing must be avoided to ensure real-time. 4) Template programming and inline functions help in performance optimization. 5) In practical applications, C can be used to implement an efficient logging system.
 How to measure thread performance in C?
Apr 28, 2025 pm 10:21 PM
How to measure thread performance in C?
Apr 28, 2025 pm 10:21 PM
Measuring thread performance in C can use the timing tools, performance analysis tools, and custom timers in the standard library. 1. Use the library to measure execution time. 2. Use gprof for performance analysis. The steps include adding the -pg option during compilation, running the program to generate a gmon.out file, and generating a performance report. 3. Use Valgrind's Callgrind module to perform more detailed analysis. The steps include running the program to generate the callgrind.out file and viewing the results using kcachegrind. 4. Custom timers can flexibly measure the execution time of a specific code segment. These methods help to fully understand thread performance and optimize code.
 How to use string streams in C?
Apr 28, 2025 pm 09:12 PM
How to use string streams in C?
Apr 28, 2025 pm 09:12 PM
The main steps and precautions for using string streams in C are as follows: 1. Create an output string stream and convert data, such as converting integers into strings. 2. Apply to serialization of complex data structures, such as converting vector into strings. 3. Pay attention to performance issues and avoid frequent use of string streams when processing large amounts of data. You can consider using the append method of std::string. 4. Pay attention to memory management and avoid frequent creation and destruction of string stream objects. You can reuse or use std::stringstream.
 An efficient way to batch insert data in MySQL
Apr 29, 2025 pm 04:18 PM
An efficient way to batch insert data in MySQL
Apr 29, 2025 pm 04:18 PM
Efficient methods for batch inserting data in MySQL include: 1. Using INSERTINTO...VALUES syntax, 2. Using LOADDATAINFILE command, 3. Using transaction processing, 4. Adjust batch size, 5. Disable indexing, 6. Using INSERTIGNORE or INSERT...ONDUPLICATEKEYUPDATE, these methods can significantly improve database operation efficiency.



 Research results:
Research results: 