 Technology peripherals
Technology peripherals
 AI
AI
 This compiler can make Python as fast as C++: up to a hundred times faster, produced by MIT
This compiler can make Python as fast as C++: up to a hundred times faster, produced by MIT
This compiler can make Python as fast as C++: up to a hundred times faster, produced by MIT
Since the rise of deep learning, Python has been one of the hottest programming languages. It dominates the fields of data science and machine learning, and even plays a starring role in scientific and mathematical computing. . Nowadays, you can find a corresponding Python package for almost any project you can imagine.
However, while the simplified syntax of a high-level language makes it easy to learn and use, it is slower than a low-level language like C or C.
Researchers at the MIT Computer Science and Artificial Intelligence Laboratory (CSAIL) hope to change that with Codon, a Python-based compiler that allows users Write Python code that runs as efficiently as C or C programs, while being customizable and adaptable to different needs and environments.
The latest paper of this research, "Codon: A Compiler for High-Performance Pythonic Applications and DSLs", was published at the 32nd ACM SIGPLAN International Conference on Compiler Construction in February.

- ## Project link: https://github.com/exaloop/codon
- Paper: https://dl.acm.org/doi/abs/10.1145/3578360.3580275
"Regular Python is compiled into so-called bytecode, which is executed in a virtual machine, which makes it much slower," said Codon, the lead author of the paper Ariya Shajii said, "With Codon, we compile natively, so you can run the final result directly on the CPU - without going through an intermediate virtual machine or interpreter."
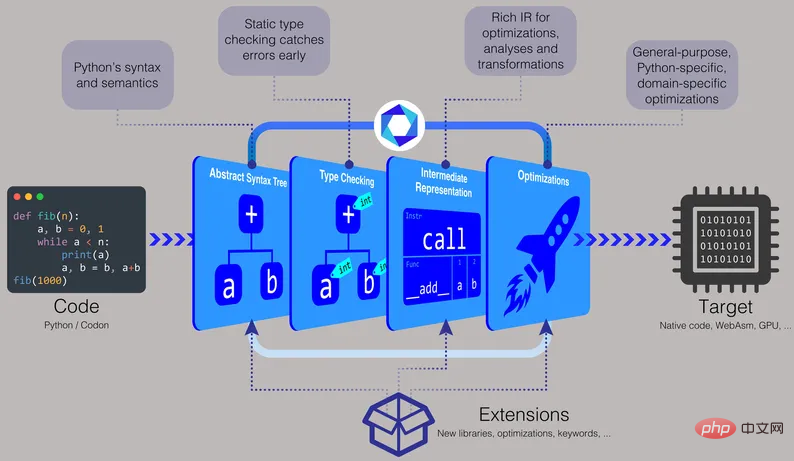
Codon's compilation pipeline includes type checking, allowing it to run Python code more efficiently.
The Python-based compiler comes with pre-built binaries for Linux and macOS, and you can also build or generate executables from source. "With Codon, you can distribute source code like Python, or you can compile it into binaries," Shajii said. "If you want to distribute a binary, it will be the same as a language like C, such as a Linux binary or a Mac binary."To make Codon faster, research People decided to perform type checking at compile time. Type checking involves assigning a data type (such as integer, string, character, or float, etc.) to a value. For example, the number 5 can be assigned as an integer, the letter c can be assigned as a character, the word hello can be assigned as a string, and the decimal number 3.14 can be assigned as a floating point number.
"In regular Python, all types are given to the runtime," Shajii said. "Using Codon, we do type checking during compilation, which allows us to avoid all expensive type manipulation at runtime."
Saman Amarasinghe, principal researcher at MIT CSAIL, added, " If you have a dynamic language (like Python), every time you have some data, you need to keep a lot of extra metadata around it to determine the type of runtime. Codon does away with this metadata, so the code is faster Faster and smaller data size."
According to Shajii, Codon does not have any unnecessary data or type checking at runtime, so there is zero overhead. In terms of performance, "Codon is generally on par with C. We typically see 10x to 100x speed improvements compared to Python."
On the other hand, Codon The approach has its trade-offs. "We do this static type checking and don't allow the use of some of Python's dynamic features, such as dynamically changing types at runtime," Shajii said.
"There are still some Python libraries that we haven't implemented yet." Amarasinghe added, "Python has been actually tested by countless people, and Codon has not reached that level yet. It needs to run more programs to obtain More feedback, and more reinforcement. It will take some time to reach a stable level of regular Python." Codon was originally designed for genomics and bioinformatics work. The researchers tried about 10 common genomics applications written in Python and compiled them using Codon, achieving speedups of 5 to 10 times compared to the original manually optimized implementation. "Data sets in these fields have become very large today, and high-level languages like Python and R are too slow to handle the terabytes of data per sequencing set," Shajii said. "That's the gap we want to fill - by building a way to process big data without having to write C or C code to help domain experts who are not computer science or professional developers." The above chart compares the performance of Python (CPython 3), PyPy, Codon, and C on several benchmarks. The y-axis shows the speedup of the Codon implementation relative to the CPython implementation. MIT/EXALOOP/UNIVERSITY OF VICTORIA/ACM In addition to genomics, Codon can also be applied to similar applications that handle massive data sets, as well as Python-based compilation GPU programming and parallel programming supported by the processor. In fact, Codon is now being used commercially in bioinformatics, deep learning, and quantitative finance through the startup Exaloop, which Shajii founded to transform Codon from an academic project into an industry application. To enable Codon to adapt to different fields, the team developed a plug-in system. "It's like an extensible compiler," Shajii said. "You can write plug-ins for genomics or other fields, and these plug-ins can have new libraries and new compiler optimizations." In addition, companies and institutions can use Codon to prototype and develop your own applications. "One of the patterns we see is that people use Python for prototyping and testing because it's easy to use, but when it comes to something important, they have to rewrite the application or have someone else do it in C or C Rewrite and test on a larger data set," Shajii said. "With Codon, you can fully use Python and get the best of both worlds." Regarding the future of Codon, Shajii and his team are currently working on native versions of the widely used Python library. implementation, as well as library-specific optimizations to help people get better performance from these libraries. They also plan to create a popular feature: Codon's WebAssembly backend to support running code on a web browser. 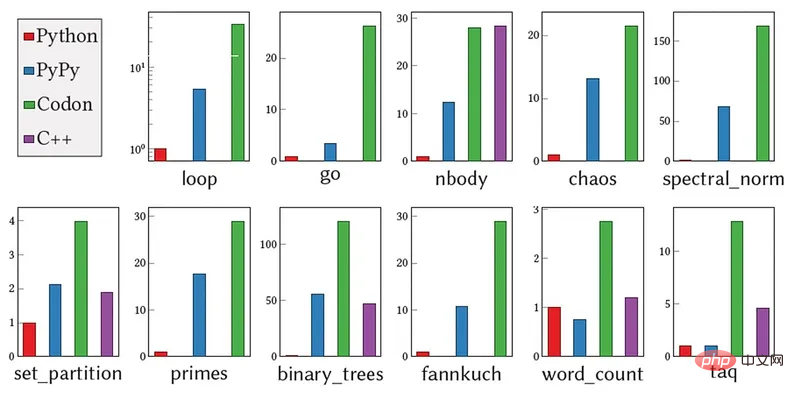
The above is the detailed content of This compiler can make Python as fast as C++: up to a hundred times faster, produced by MIT. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

AI Hentai Generator
Generate AI Hentai for free.

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1378
1378
 52
52
 Python: Games, GUIs, and More
Apr 13, 2025 am 12:14 AM
Python: Games, GUIs, and More
Apr 13, 2025 am 12:14 AM
Python excels in gaming and GUI development. 1) Game development uses Pygame, providing drawing, audio and other functions, which are suitable for creating 2D games. 2) GUI development can choose Tkinter or PyQt. Tkinter is simple and easy to use, PyQt has rich functions and is suitable for professional development.
 PHP and Python: Comparing Two Popular Programming Languages
Apr 14, 2025 am 12:13 AM
PHP and Python: Comparing Two Popular Programming Languages
Apr 14, 2025 am 12:13 AM
PHP and Python each have their own advantages, and choose according to project requirements. 1.PHP is suitable for web development, especially for rapid development and maintenance of websites. 2. Python is suitable for data science, machine learning and artificial intelligence, with concise syntax and suitable for beginners.
 How debian readdir integrates with other tools
Apr 13, 2025 am 09:42 AM
How debian readdir integrates with other tools
Apr 13, 2025 am 09:42 AM
The readdir function in the Debian system is a system call used to read directory contents and is often used in C programming. This article will explain how to integrate readdir with other tools to enhance its functionality. Method 1: Combining C language program and pipeline First, write a C program to call the readdir function and output the result: #include#include#include#includeintmain(intargc,char*argv[]){DIR*dir;structdirent*entry;if(argc!=2){
 Python and Time: Making the Most of Your Study Time
Apr 14, 2025 am 12:02 AM
Python and Time: Making the Most of Your Study Time
Apr 14, 2025 am 12:02 AM
To maximize the efficiency of learning Python in a limited time, you can use Python's datetime, time, and schedule modules. 1. The datetime module is used to record and plan learning time. 2. The time module helps to set study and rest time. 3. The schedule module automatically arranges weekly learning tasks.
 Nginx SSL Certificate Update Debian Tutorial
Apr 13, 2025 am 07:21 AM
Nginx SSL Certificate Update Debian Tutorial
Apr 13, 2025 am 07:21 AM
This article will guide you on how to update your NginxSSL certificate on your Debian system. Step 1: Install Certbot First, make sure your system has certbot and python3-certbot-nginx packages installed. If not installed, please execute the following command: sudoapt-getupdatesudoapt-getinstallcertbotpython3-certbot-nginx Step 2: Obtain and configure the certificate Use the certbot command to obtain the Let'sEncrypt certificate and configure Nginx: sudocertbot--nginx Follow the prompts to select
 GitLab's plug-in development guide on Debian
Apr 13, 2025 am 08:24 AM
GitLab's plug-in development guide on Debian
Apr 13, 2025 am 08:24 AM
Developing a GitLab plugin on Debian requires some specific steps and knowledge. Here is a basic guide to help you get started with this process. Installing GitLab First, you need to install GitLab on your Debian system. You can refer to the official installation manual of GitLab. Get API access token Before performing API integration, you need to get GitLab's API access token first. Open the GitLab dashboard, find the "AccessTokens" option in the user settings, and generate a new access token. Will be generated
 How to configure HTTPS server in Debian OpenSSL
Apr 13, 2025 am 11:03 AM
How to configure HTTPS server in Debian OpenSSL
Apr 13, 2025 am 11:03 AM
Configuring an HTTPS server on a Debian system involves several steps, including installing the necessary software, generating an SSL certificate, and configuring a web server (such as Apache or Nginx) to use an SSL certificate. Here is a basic guide, assuming you are using an ApacheWeb server. 1. Install the necessary software First, make sure your system is up to date and install Apache and OpenSSL: sudoaptupdatesudoaptupgradesudoaptinsta
 What service is apache
Apr 13, 2025 pm 12:06 PM
What service is apache
Apr 13, 2025 pm 12:06 PM
Apache is the hero behind the Internet. It is not only a web server, but also a powerful platform that supports huge traffic and provides dynamic content. It provides extremely high flexibility through a modular design, allowing for the expansion of various functions as needed. However, modularity also presents configuration and performance challenges that require careful management. Apache is suitable for server scenarios that require highly customizable and meet complex needs.



