 Technology peripherals
Technology peripherals
 AI
AI
 Faster than 0! Meta launched a large protein model with 15 billion parameters to crush AlphaFold2
Faster than 0! Meta launched a large protein model with 15 billion parameters to crush AlphaFold2
Faster than 0! Meta launched a large protein model with 15 billion parameters to crush AlphaFold2
The largest protein language model to date has been released!
A year ago, DeepMind’s open source AlphaFold2 was launched in Nature and Science, overwhelming the biological and AI academic circles.
A year later, Meta came with ESMFold, which was an order of magnitude faster.
Not only is it fast, the model also has 15 billion parameters.
LeCun tweeted to praise this as a great new achievement by the Meta-FAIR protein team.

Co-author Zeming Lin revealed that the large model with 3 billion parameters was trained on 256 GPUs for 3 weeks, while ESMfold took 10 days on 128 GPUs. As for the 15 billion parameter version, it is still unclear.
He also said that the code will definitely be open sourced later, so stay tuned!
Big and fast!
Today, our protagonist is ESMFold, a model that directly predicts high-accuracy, end-to-end, atomic-level structure from individual protein sequences.

Paper address: https://www.biorxiv.org/content/10.1101/2022.07.20.500902v1
The benefits of 15 billion parameters Needless to say – today’s large models can be trained to predict the three-dimensional structure of proteins with atomic-sized accuracy.
In terms of accuracy, ESMFold is similar to AlphaFold2 and RoseTTAFold.
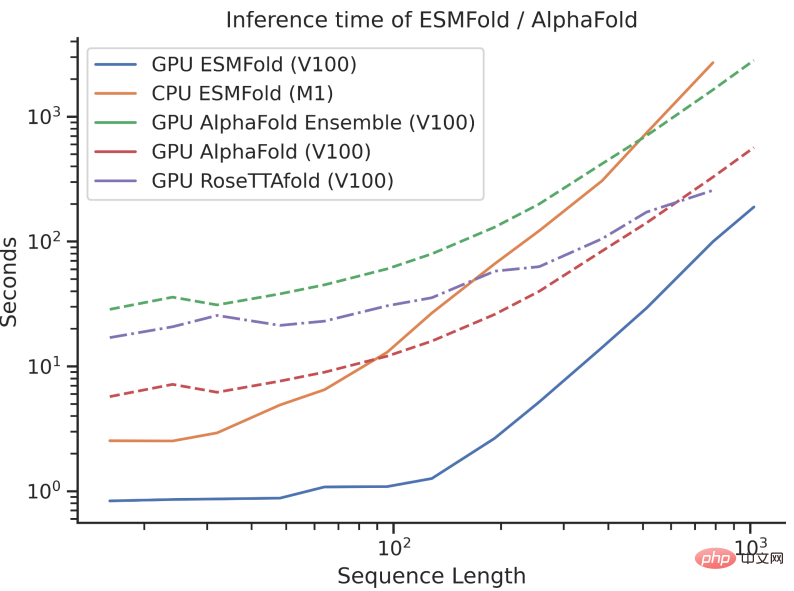
However, ESMFold’s inference speed is an order of magnitude faster than AlphaFold2!
It may be difficult to understand the speed comparison between the three by talking about the order of magnitude. Just look at the picture below to understand.
What’s the difference?
Although AlphaFold2 and RoseTTAFold have achieved breakthrough success in the problem of atomic resolution structure prediction, they also rely on the use of multiple sequence alignments (MSA) and similar protein structure templates for optimal performance.
In contrast, by leveraging the internal representation of the language model, ESMFold can generate corresponding structure predictions using only one sequence as input, thus greatly speeding up structure prediction.
The researchers found that ESMFold’s predictions for low-complexity sequences were comparable to current state-of-the-art models.
Moreover, the accuracy of structure prediction is closely related to the complexity of the language model. That is to say, when the language model can better understand the sequence, it can better understand the structure.
Currently, there are billions of protein sequences of unknown structure and function, many of which are derived from metagenomic sequencing.
Using ESMFold, researchers can fold a random sample of 1 million metagenomic sequences in just 6 hours.
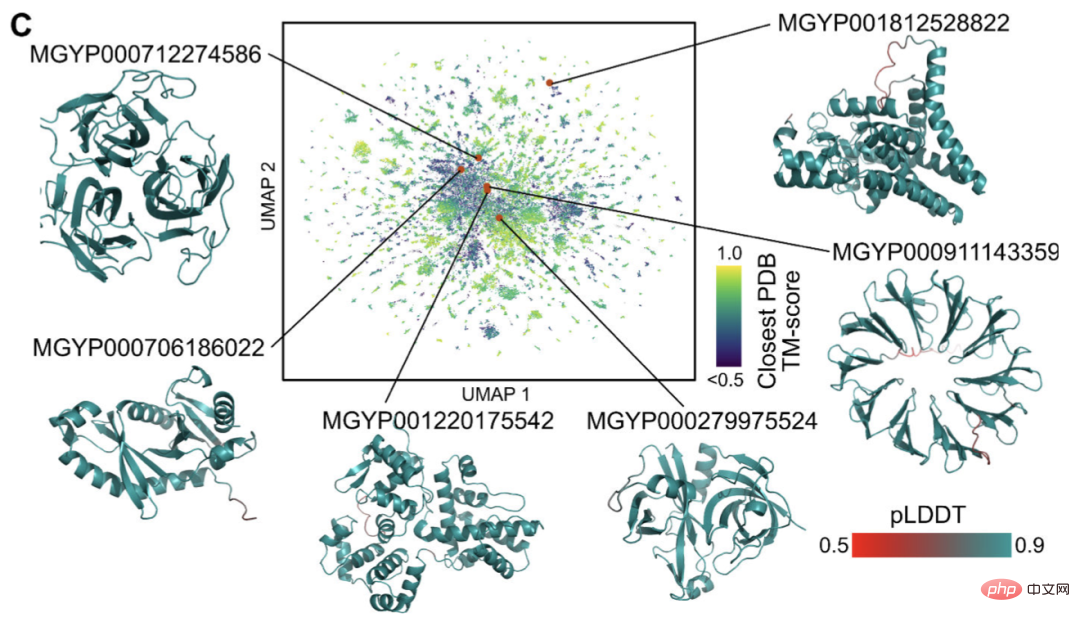
A large proportion of these have high confidence and are unlike any known structure (have no records in the database).
Researchers believe that ESMFold can help understand protein structures that are beyond current understanding.
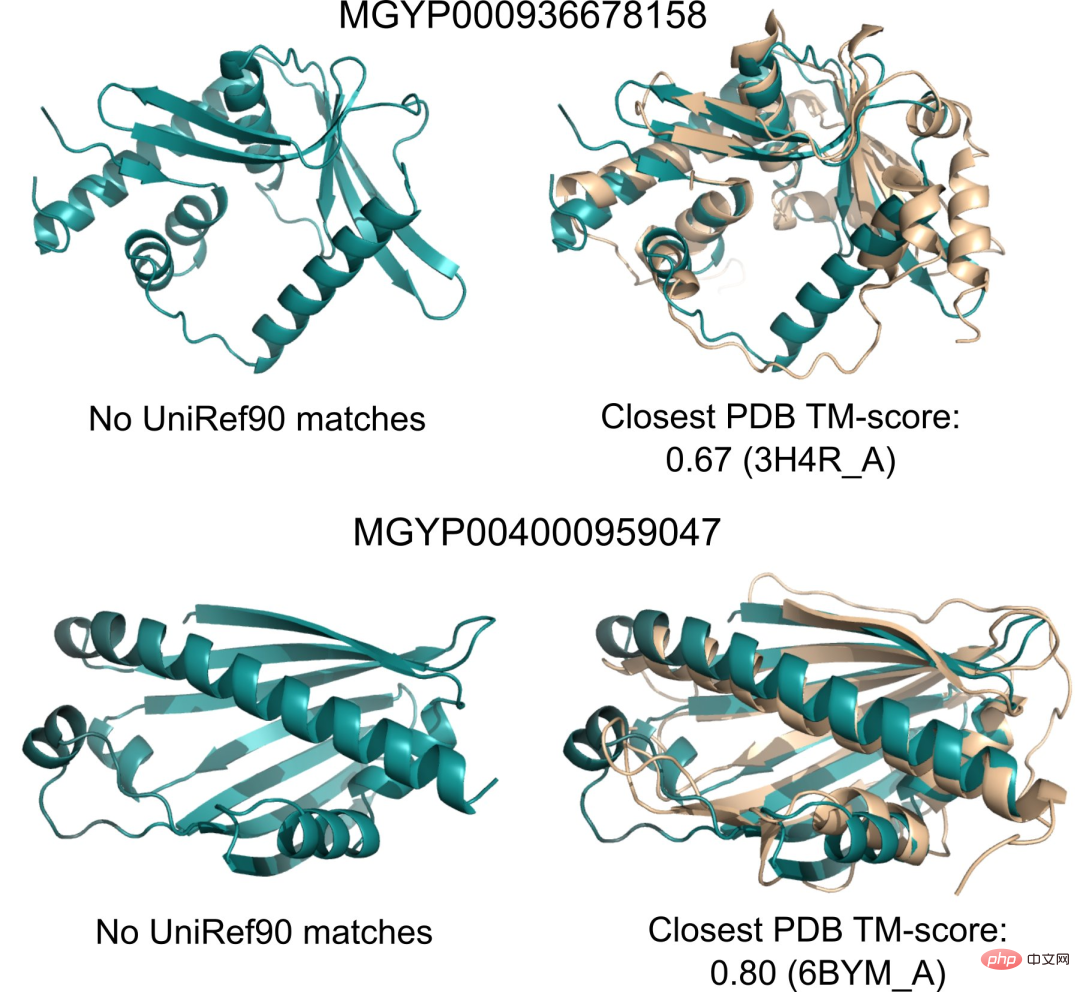
Additionally, because ESMFold’s predictions are an order of magnitude faster than existing models, researchers can use ESMFold to help fill rapidly growing protein sequence databases and slow progress. The gap between protein structure and function databases.
15 billion parameter protein language model
Next let’s talk about Meta’s new ESMFold in detail.
ESM-2 is a Transformer-based language model and uses an attention mechanism to learn the interaction patterns between pairs of amino acids in the input sequence.
Compared with the previous generation model ESM-1b, Meta has improved the model structure and training parameters, and added computing resources and data. At the same time, the addition of relative position embedding enables the model to be generalized to sequences of any length.
From the results, the ESM-2 model with 150 million parameters performed better than the ESM-1b model with 650 million parameters.
In addition, ESM-2 also surpasses other protein language models on the benchmark of structure prediction. This performance improvement is consistent with established patterns in the large language modeling field.
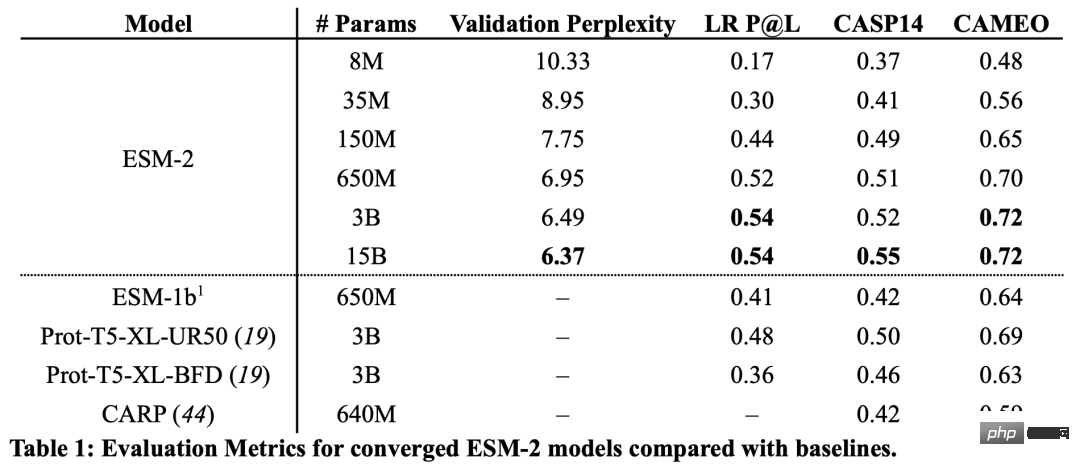
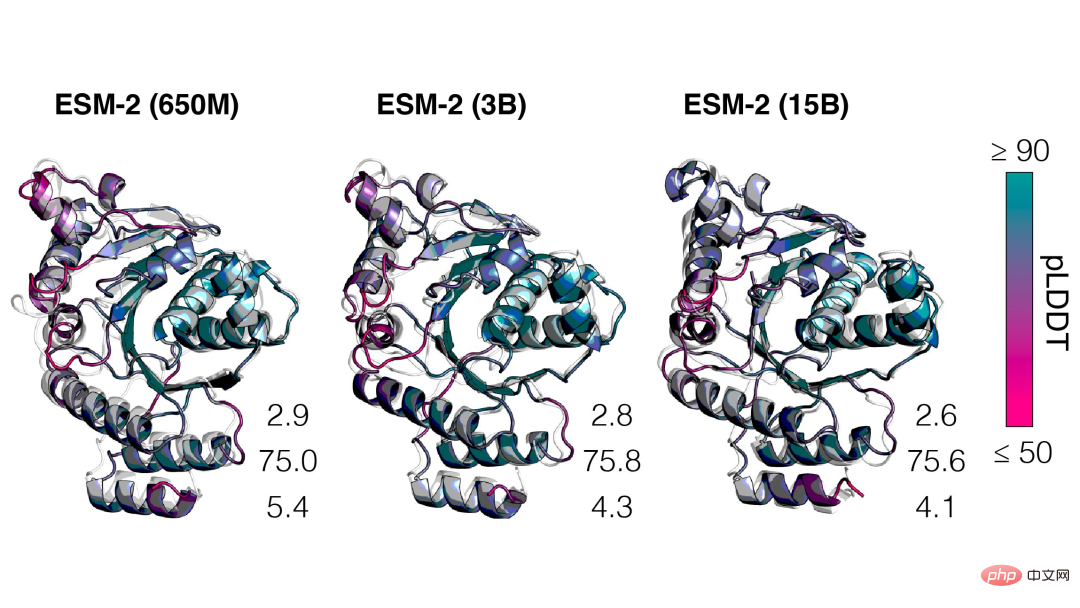
As the scale of ESM-2 increases, it can be observed that the accuracy of language modeling has greatly improved.
End-to-end single sequence structure prediction
A key difference between SMFold and AlphaFold2 is that ESMFold uses language model representation, which eliminates the need for explicit homology Sequences (in the form of MSA) are required as input.
ESMFold simplifies the Evoformer in AlphaFold2 by replacing the computationally expensive network module that handles MSA with a Transformer module that handles sequences. This simplification means that ESMFold is significantly faster than MSA-based models.
The output of the folded backbone is then processed by a structure module, which is responsible for outputting the final atomic-level structure and prediction confidence.
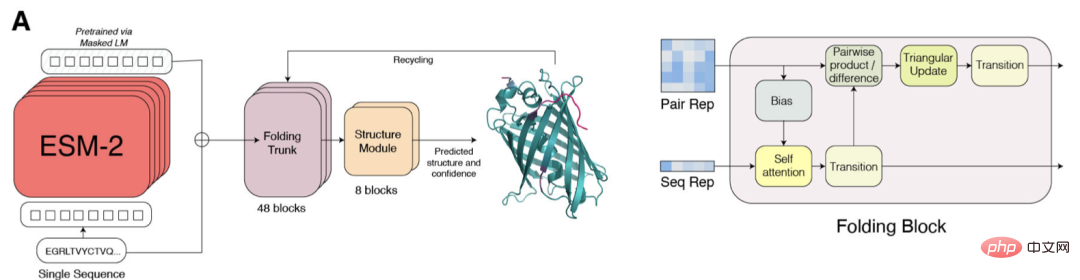
Researchers compared ESMFold with AlphaFold2 and RoseTTAFold on the CAMEO (April 2022 to June 2022) and CASP14 (May 2020) test sets.
When only a single sequence is given as input, ESMFold performs much better than Alphafold 2.
When using the complete pipeline, AlphaFold2 achieved 88.3 and 84.7 on CAMEO and CASP14 respectively. ESMFold achieves comparable accuracy to RoseTTAfold on CAMEO, with an average TM score of 82.0.
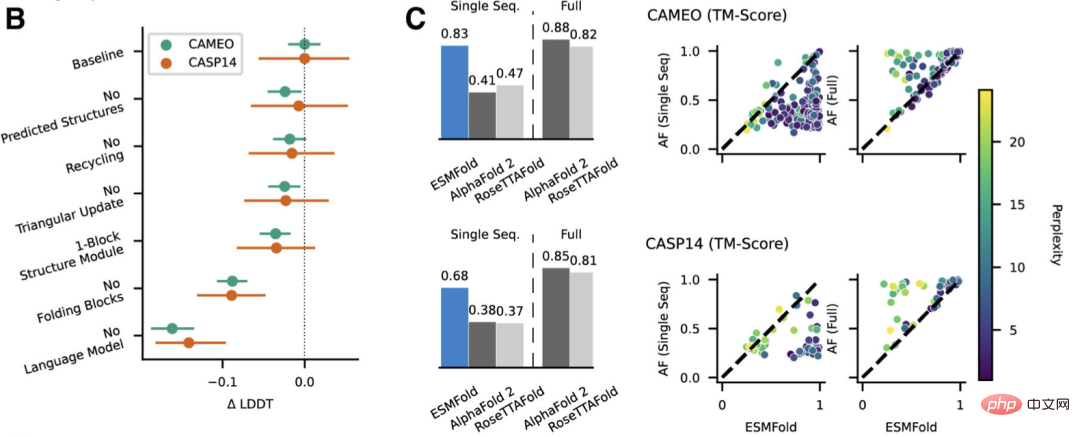
Conclusion
The researchers found that language models targeting unsupervised learning performed well on a large Trained on an evolutionarily diverse protein sequence database, it can predict protein structures with atomic-level resolution.
By expanding the parameters of the language model to 15B, the impact of scale on protein structure learning can be systematically studied.
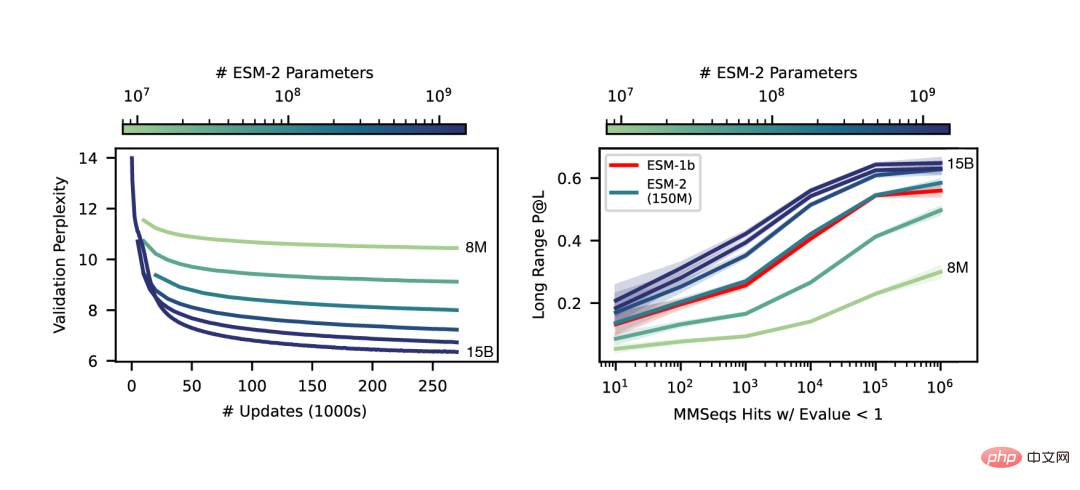
We saw that the nonlinear curve of protein structure predictions is a function of model size, and observed a strong connection between how well a language model understands a sequence and its structure predictions.
The models of the ESM-2 series are the largest protein language models trained to date, with only an order of magnitude fewer parameters than the largest recently developed text models.
Moreover, ESM-2 is a very big improvement over the previous model. Even under 150M parameters, ESM-2 captures more accurately than the ESM-1 generation language model under 650 million parameters. Structure diagram.
Researchers said that the biggest driver of ESMFold performance is the language model. Because there is a strong connection between the perplexity of language models and the accuracy of structure predictions, they found that when ESM-2 can better understand protein sequences, it can achieve predictions comparable to current state-of-the-art models.
ESMFold has obtained accurate atomic resolution structure prediction, and the inference time is an order of magnitude faster than AlphaFold2.
In practice, the speed advantage is even greater. Because ESMFold does not need to search for evolutionarily related sequences to construct MSA.
Although there are faster ways to reduce search time, it may still be very long no matter how much it is reduced.
The benefits brought by the greatly shortened inference time are self-evident - the increase in speed will make it possible to map the structural space of large metagenomics sequence databases.
In addition to structure-based tools to identify distal homology and conservation, rapid and accurate structure prediction with ESMFold can also play an important role in the structural and functional analysis of large new sequence collections.
Obtaining millions of predicted structures in a limited time will help discover new understanding of the breadth and diversity of natural proteins and enable the discovery of completely new protein structures and protein functions.
Introduction to the author
The co-author of this article is Zeming Lin from Meta AI.

According to his personal homepage, Zeming studied for a PhD at New York University and worked as a research engineer (visiting) at Meta AI, mainly responsible for back-end infrastructure work.
He studied at the University of Virginia for both his bachelor's and master's degrees, where he and Yanjun Qi did research on machine learning applications, especially in protein structure prediction.
The areas of interest are deep learning, structure prediction, and information biology.
The above is the detailed content of Faster than 0! Meta launched a large protein model with 15 billion parameters to crush AlphaFold2. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

AI Hentai Generator
Generate AI Hentai for free.

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1377
1377
 52
52
 DeepMind robot plays table tennis, and its forehand and backhand slip into the air, completely defeating human beginners
Aug 09, 2024 pm 04:01 PM
DeepMind robot plays table tennis, and its forehand and backhand slip into the air, completely defeating human beginners
Aug 09, 2024 pm 04:01 PM
But maybe he can’t defeat the old man in the park? The Paris Olympic Games are in full swing, and table tennis has attracted much attention. At the same time, robots have also made new breakthroughs in playing table tennis. Just now, DeepMind proposed the first learning robot agent that can reach the level of human amateur players in competitive table tennis. Paper address: https://arxiv.org/pdf/2408.03906 How good is the DeepMind robot at playing table tennis? Probably on par with human amateur players: both forehand and backhand: the opponent uses a variety of playing styles, and the robot can also withstand: receiving serves with different spins: However, the intensity of the game does not seem to be as intense as the old man in the park. For robots, table tennis
 Understand Tokenization in one article!
Apr 12, 2024 pm 02:31 PM
Understand Tokenization in one article!
Apr 12, 2024 pm 02:31 PM
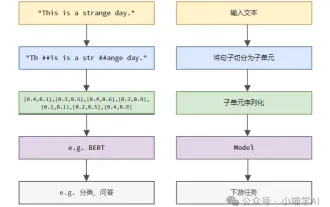
Language models reason about text, which is usually in the form of strings, but the input to the model can only be numbers, so the text needs to be converted into numerical form. Tokenization is a basic task of natural language processing. It can divide a continuous text sequence (such as sentences, paragraphs, etc.) into a character sequence (such as words, phrases, characters, punctuation, etc.) according to specific needs. The units in it Called a token or word. According to the specific process shown in the figure below, the text sentences are first divided into units, then the single elements are digitized (mapped into vectors), then these vectors are input to the model for encoding, and finally output to downstream tasks to further obtain the final result. Text segmentation can be divided into Toke according to the granularity of text segmentation.
 Efficient parameter fine-tuning of large-scale language models--BitFit/Prefix/Prompt fine-tuning series
Oct 07, 2023 pm 12:13 PM
Efficient parameter fine-tuning of large-scale language models--BitFit/Prefix/Prompt fine-tuning series
Oct 07, 2023 pm 12:13 PM
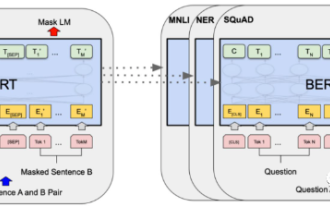
In 2018, Google released BERT. Once it was released, it defeated the State-of-the-art (Sota) results of 11 NLP tasks in one fell swoop, becoming a new milestone in the NLP world. The structure of BERT is shown in the figure below. On the left is the BERT model preset. The training process, on the right is the fine-tuning process for specific tasks. Among them, the fine-tuning stage is for fine-tuning when it is subsequently used in some downstream tasks, such as text classification, part-of-speech tagging, question and answer systems, etc. BERT can be fine-tuned on different tasks without adjusting the structure. Through the task design of "pre-trained language model + downstream task fine-tuning", it brings powerful model effects. Since then, "pre-training language model + downstream task fine-tuning" has become the mainstream training in the NLP field.
 To provide a new scientific and complex question answering benchmark and evaluation system for large models, UNSW, Argonne, University of Chicago and other institutions jointly launched the SciQAG framework
Jul 25, 2024 am 06:42 AM
To provide a new scientific and complex question answering benchmark and evaluation system for large models, UNSW, Argonne, University of Chicago and other institutions jointly launched the SciQAG framework
Jul 25, 2024 am 06:42 AM
Editor |ScienceAI Question Answering (QA) data set plays a vital role in promoting natural language processing (NLP) research. High-quality QA data sets can not only be used to fine-tune models, but also effectively evaluate the capabilities of large language models (LLM), especially the ability to understand and reason about scientific knowledge. Although there are currently many scientific QA data sets covering medicine, chemistry, biology and other fields, these data sets still have some shortcomings. First, the data form is relatively simple, most of which are multiple-choice questions. They are easy to evaluate, but limit the model's answer selection range and cannot fully test the model's ability to answer scientific questions. In contrast, open-ended Q&A
 Three secrets for deploying large models in the cloud
Apr 24, 2024 pm 03:00 PM
Three secrets for deploying large models in the cloud
Apr 24, 2024 pm 03:00 PM
Compilation|Produced by Xingxuan|51CTO Technology Stack (WeChat ID: blog51cto) In the past two years, I have been more involved in generative AI projects using large language models (LLMs) rather than traditional systems. I'm starting to miss serverless cloud computing. Their applications range from enhancing conversational AI to providing complex analytics solutions for various industries, and many other capabilities. Many enterprises deploy these models on cloud platforms because public cloud providers already provide a ready-made ecosystem and it is the path of least resistance. However, it doesn't come cheap. The cloud also offers other benefits such as scalability, efficiency and advanced computing capabilities (GPUs available on demand). There are some little-known aspects of deploying LLM on public cloud platforms
 AlphaFold 3 is launched, comprehensively predicting the interactions and structures of proteins and all living molecules, with far greater accuracy than ever before
Jul 16, 2024 am 12:08 AM
AlphaFold 3 is launched, comprehensively predicting the interactions and structures of proteins and all living molecules, with far greater accuracy than ever before
Jul 16, 2024 am 12:08 AM
Editor | Radish Skin Since the release of the powerful AlphaFold2 in 2021, scientists have been using protein structure prediction models to map various protein structures within cells, discover drugs, and draw a "cosmic map" of every known protein interaction. . Just now, Google DeepMind released the AlphaFold3 model, which can perform joint structure predictions for complexes including proteins, nucleic acids, small molecules, ions and modified residues. The accuracy of AlphaFold3 has been significantly improved compared to many dedicated tools in the past (protein-ligand interaction, protein-nucleic acid interaction, antibody-antigen prediction). This shows that within a single unified deep learning framework, it is possible to achieve
 RoSA: A new method for efficient fine-tuning of large model parameters
Jan 18, 2024 pm 05:27 PM
RoSA: A new method for efficient fine-tuning of large model parameters
Jan 18, 2024 pm 05:27 PM
As language models scale to unprecedented scale, comprehensive fine-tuning for downstream tasks becomes prohibitively expensive. In order to solve this problem, researchers began to pay attention to and adopt the PEFT method. The main idea of the PEFT method is to limit the scope of fine-tuning to a small set of parameters to reduce computational costs while still achieving state-of-the-art performance on natural language understanding tasks. In this way, researchers can save computing resources while maintaining high performance, bringing new research hotspots to the field of natural language processing. RoSA is a new PEFT technique that, through experiments on a set of benchmarks, is found to outperform previous low-rank adaptive (LoRA) and pure sparse fine-tuning methods using the same parameter budget. This article will go into depth
 Conveniently trained the biggest ViT in history? Google upgrades visual language model PaLI: supports 100+ languages
Apr 12, 2023 am 09:31 AM
Conveniently trained the biggest ViT in history? Google upgrades visual language model PaLI: supports 100+ languages
Apr 12, 2023 am 09:31 AM
The progress of natural language processing in recent years has largely come from large-scale language models. Each new model released pushes the amount of parameters and training data to new highs, and at the same time, the existing benchmark rankings will be slaughtered. ! For example, in April this year, Google released the 540 billion-parameter language model PaLM (Pathways Language Model), which successfully surpassed humans in a series of language and reasoning tests, especially its excellent performance in few-shot small sample learning scenarios. PaLM is considered to be the development direction of the next generation language model. In the same way, visual language models actually work wonders, and performance can be improved by increasing the scale of the model. Of course, if it is just a multi-tasking visual language model



