 Technology peripherals
Technology peripherals
 AI
AI
 Endless Possibilities NVIDIA Generative AI Model Speeds Protein Synthesis
Endless Possibilities NVIDIA Generative AI Model Speeds Protein Synthesis
Endless Possibilities NVIDIA Generative AI Model Speeds Protein Synthesis
Over the past two years, machine learning has revolutionized protein structure prediction. Now, artificial intelligence has triggered a new round of revolution in the field of protein design.
Since the advent of AI, many scientists have joined the track of using it to conduct protein research. Biologists have discovered that using machine learning, protein molecules can be created in seconds. In the past, this time might have been several months.
Recently, the start-up Evozyne used pre-trained AI models provided by NVIDIA to create two proteins with significant potential in the medical and clean energy fields. One of the proteins is used to treat a congenital disease, and another is used to consume carbon dioxide to reduce global warming.
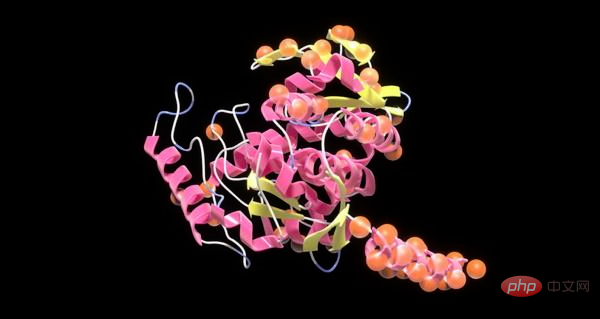
So scientists use NVIDIA BioNeMo to create large language models that generate high-quality proteins to speed up drug development and help create a more sustainable environment.
New ways to accelerate drug discovery
Andrew Ferguson, co-founder of Evozyne and co-author of the paper, said: “It is gratifying that this AI model has produced the first round of production. The resulting synthetic proteins are just like naturally occurring proteins, indicating that the model has learned nature's design rules."
Evozyne uses NVIDIA's ProtT5. ProtT5 is a Transformer model that is part of NVIDIA BioNeMo, a software framework and service for creating medical AI models.
Molecular engineer Ferguson, whose research fields include chemistry and machine learning, said: "BioNeMo is very powerful and allows us to train a model and then use the model to run work tasks at a very low cost, in a matter of seconds. Millions of sequences can be generated in minutes. The model predicts how to assemble new proteins that meet Evozyne's needs."
The model is at the heart of the Evovyne ProT-VAE pipeline. Evozyne’s ProT-VAE pipeline combines the powerful Transformer model in NVIDIA BioNeMo with variational autoencoders (VAEs).
He said: "A few years ago, no one noticed that you could use large language models combined with variational autoencoders to design proteins."
In contrast, Evozyne's method can change half or more of the amino acids in a protein in just one round. This amounts to hundreds of mutations.
Evozyne data scientist Joshua Moller said: "They speed up training by extending the work to multiple GPUs.
This reduces the time to train large AI models from months to a week "So we can train models that would otherwise be impossible to train," Ferguson said, "such as models with billions of trainable parameters. ”
Revolutionary AI Model
The traditional protein engineering design method, namely directed evolution, uses a slow, unplanned approach, usually just one protein at a time. Change the sequence of a few amino acids. Machine learning helps to study a large number of possible amino acid combinations and then effectively identify the most useful sequences.
BioNeMo is an AI-enabled drug development built on NVIDIA NeMo Megatron Cloud service and framework for training and deploying large biomolecule Transformer AI models at supercomputing scale. Services include pre-trained LLM, native support for common file formats for proteins, DNA, RNA and chemistry, and also provides support for SMILES ( Data loader used by (for molecular structures) and FASTA (for amino acid and nucleotide sequences).
With BioNeMo, scientists can start easily using pre-trained models, automatic downloaders and Preprocessor. With unsupervised structured learners, various models, embeddings, and outputs are combined to combine multimodal data. Unsupervised pretraining also eliminates the need for labeled data, allowing for rapid generation Learned embeddings predict protein structure, function, cellular location, water solubility, membrane binding, conservation and variable regions, etc.
Among them, MegaMolBART is a method that uses 1.4 billion molecules (SMILES string ), which can be used for a variety of chemical informatics applications. Moreover, BioNeMo provides Transformer-based protein language models such as ProtT5 and ESM1-85M.
BioNeMo also provides OpenFold, which is a user-friendly A deep learning model for predicting the 3D structure of novel protein sequences.
NVIDIA's Transformer model reads the amino acid sequences in millions of proteins. The model uses the technology used by neural networks to understand text and learns how to How nature builds protein amino acid sequences.
Looking to the future, the prospects of using AI to accelerate protein engineering are very broad. Artificially designed proteins are more stable than proteins that originally exist in nature and can survive without energy or One of its functions can also be realized under extreme conditions such as high temperature.
In addition, artificial intelligence can also be used to design amino acid sequences to match the backbone, which can be used to improve the stability of specific proteins such as enzymes and antibodies. Artificial intelligence technology plays a very important role in the design of proteins of different sizes and different conformations. In the future, it can also help design more and more useful proteins, including new biological materials that can be used to reduce pollution and improve the environment.
The above is the detailed content of Endless Possibilities NVIDIA Generative AI Model Speeds Protein Synthesis. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

AI Hentai Generator
Generate AI Hentai for free.

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1386
1386
 52
52
 Centos shutdown command line
Apr 14, 2025 pm 09:12 PM
Centos shutdown command line
Apr 14, 2025 pm 09:12 PM
The CentOS shutdown command is shutdown, and the syntax is shutdown [Options] Time [Information]. Options include: -h Stop the system immediately; -P Turn off the power after shutdown; -r restart; -t Waiting time. Times can be specified as immediate (now), minutes ( minutes), or a specific time (hh:mm). Added information can be displayed in system messages.
 How to check CentOS HDFS configuration
Apr 14, 2025 pm 07:21 PM
How to check CentOS HDFS configuration
Apr 14, 2025 pm 07:21 PM
Complete Guide to Checking HDFS Configuration in CentOS Systems This article will guide you how to effectively check the configuration and running status of HDFS on CentOS systems. The following steps will help you fully understand the setup and operation of HDFS. Verify Hadoop environment variable: First, make sure the Hadoop environment variable is set correctly. In the terminal, execute the following command to verify that Hadoop is installed and configured correctly: hadoopversion Check HDFS configuration file: The core configuration file of HDFS is located in the /etc/hadoop/conf/ directory, where core-site.xml and hdfs-site.xml are crucial. use
 What are the backup methods for GitLab on CentOS
Apr 14, 2025 pm 05:33 PM
What are the backup methods for GitLab on CentOS
Apr 14, 2025 pm 05:33 PM
Backup and Recovery Policy of GitLab under CentOS System In order to ensure data security and recoverability, GitLab on CentOS provides a variety of backup methods. This article will introduce several common backup methods, configuration parameters and recovery processes in detail to help you establish a complete GitLab backup and recovery strategy. 1. Manual backup Use the gitlab-rakegitlab:backup:create command to execute manual backup. This command backs up key information such as GitLab repository, database, users, user groups, keys, and permissions. The default backup file is stored in the /var/opt/gitlab/backups directory. You can modify /etc/gitlab
 Centos install mysql
Apr 14, 2025 pm 08:09 PM
Centos install mysql
Apr 14, 2025 pm 08:09 PM
Installing MySQL on CentOS involves the following steps: Adding the appropriate MySQL yum source. Execute the yum install mysql-server command to install the MySQL server. Use the mysql_secure_installation command to make security settings, such as setting the root user password. Customize the MySQL configuration file as needed. Tune MySQL parameters and optimize databases for performance.
 How is the GPU support for PyTorch on CentOS
Apr 14, 2025 pm 06:48 PM
How is the GPU support for PyTorch on CentOS
Apr 14, 2025 pm 06:48 PM
Enable PyTorch GPU acceleration on CentOS system requires the installation of CUDA, cuDNN and GPU versions of PyTorch. The following steps will guide you through the process: CUDA and cuDNN installation determine CUDA version compatibility: Use the nvidia-smi command to view the CUDA version supported by your NVIDIA graphics card. For example, your MX450 graphics card may support CUDA11.1 or higher. Download and install CUDAToolkit: Visit the official website of NVIDIACUDAToolkit and download and install the corresponding version according to the highest CUDA version supported by your graphics card. Install cuDNN library:
 Detailed explanation of docker principle
Apr 14, 2025 pm 11:57 PM
Detailed explanation of docker principle
Apr 14, 2025 pm 11:57 PM
Docker uses Linux kernel features to provide an efficient and isolated application running environment. Its working principle is as follows: 1. The mirror is used as a read-only template, which contains everything you need to run the application; 2. The Union File System (UnionFS) stacks multiple file systems, only storing the differences, saving space and speeding up; 3. The daemon manages the mirrors and containers, and the client uses them for interaction; 4. Namespaces and cgroups implement container isolation and resource limitations; 5. Multiple network modes support container interconnection. Only by understanding these core concepts can you better utilize Docker.
 How to choose a GitLab database in CentOS
Apr 14, 2025 pm 05:39 PM
How to choose a GitLab database in CentOS
Apr 14, 2025 pm 05:39 PM
When installing and configuring GitLab on a CentOS system, the choice of database is crucial. GitLab is compatible with multiple databases, but PostgreSQL and MySQL (or MariaDB) are most commonly used. This article analyzes database selection factors and provides detailed installation and configuration steps. Database Selection Guide When choosing a database, you need to consider the following factors: PostgreSQL: GitLab's default database is powerful, has high scalability, supports complex queries and transaction processing, and is suitable for large application scenarios. MySQL/MariaDB: a popular relational database widely used in Web applications, with stable and reliable performance. MongoDB:NoSQL database, specializes in
 How to view GitLab logs under CentOS
Apr 14, 2025 pm 06:18 PM
How to view GitLab logs under CentOS
Apr 14, 2025 pm 06:18 PM
A complete guide to viewing GitLab logs under CentOS system This article will guide you how to view various GitLab logs in CentOS system, including main logs, exception logs, and other related logs. Please note that the log file path may vary depending on the GitLab version and installation method. If the following path does not exist, please check the GitLab installation directory and configuration files. 1. View the main GitLab log Use the following command to view the main log file of the GitLabRails application: Command: sudocat/var/log/gitlab/gitlab-rails/production.log This command will display product



