 Technology peripherals
Technology peripherals
 AI
AI
 Meta lets a 15 billion parameter language model learn to design 'new' proteins from scratch! LeCun: Amazing results
Meta lets a 15 billion parameter language model learn to design 'new' proteins from scratch! LeCun: Amazing results
Meta lets a 15 billion parameter language model learn to design 'new' proteins from scratch! LeCun: Amazing results
AI has once again made new progress in the field of biomedicine. Yes, this time it’s about protein.
The difference is that in the past, AI discovered protein structures, but this time it began to design and generate protein structures on its own. If he was a "Prosecutor" in the past, it is not impossible to say that he has evolved into a "Creator" now.
# Participants in this study are the protein research team of FAIR, which is part of Meta’s AI research institute. As the chief AI scientist who has worked at Facebook for many years, Yann LeCun also immediately forwarded the results of his team and spoke highly of it.

These two papers on BioRxiv are Meta’s “amazing” achievements in protein design/generation. The system uses a simulated annealing algorithm to find an amino acid sequence that folds into a desired shape or satisfies constraints such as symmetry.
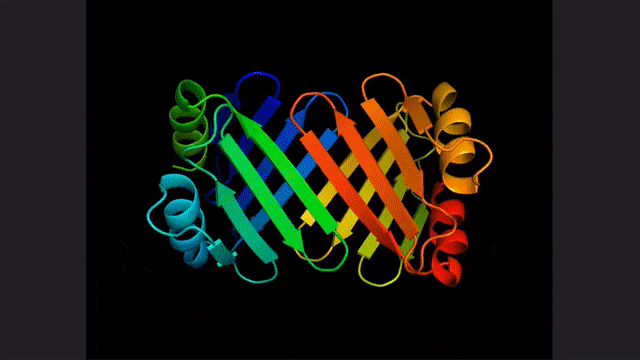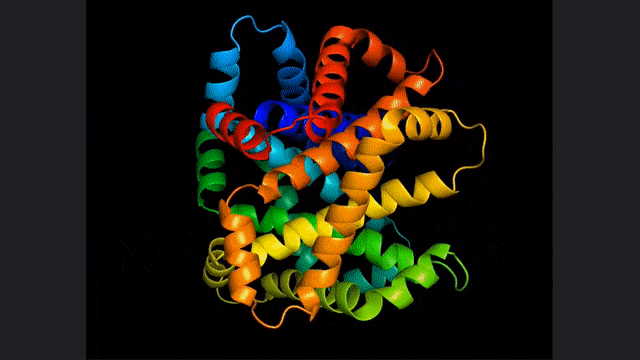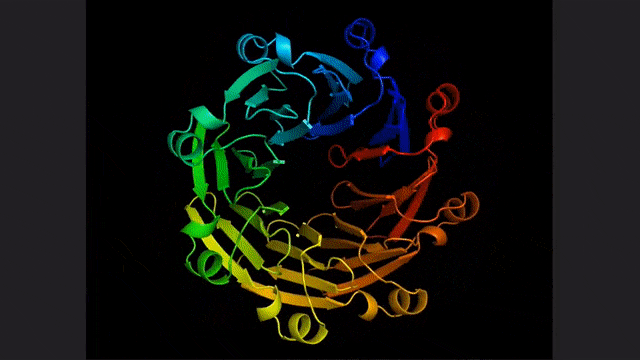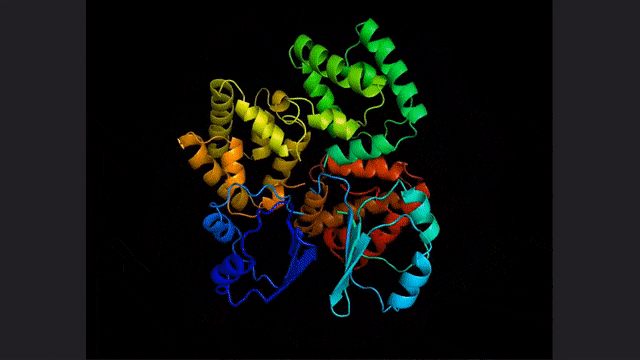
ESM2, a model for predicting atomic hierarchical structure
You guessed it right, this study is the same as the two papers The basis is the large language model for protein prediction and discovery proposed by Meta not long ago: ESM2.
This is a large model with 15 billion parameters. As the model scales from 8 to 15 million parameters, information emerging from the internal representation enables three-dimensional structure predictions at atomic resolution.
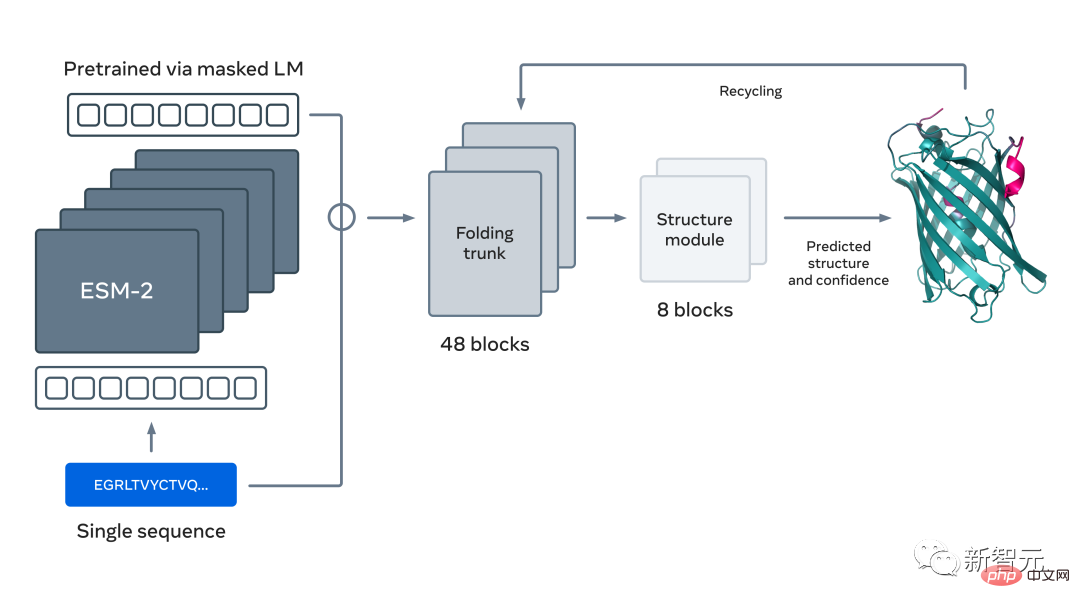
Utilizing large language models to learn evolutionary patterns, accurate structures can be generated end-to-end directly from protein sequences Predictions, while maintaining accuracy, are up to 60 times faster than current state-of-the-art methods.
#In fact, with the help of this new structural prediction capability, Meta was able to use a cluster of approximately 2,000 GPUs in just two weeks. The sequences of more than 600 million metagenomic proteins in the map were predicted.
The corresponding author of the two papers, Alex Rives from Meta AI, said that the versatility demonstrated by the ESM2 language model not only extends beyond the scope of natural proteins, but also It also enables programmable generation of complex and modular protein structures.
Protein Design "Specialized Programming Language"
If a worker wants to do his job well, he must first sharpen his tools.
In order to make protein design and generation more efficient, researchers have also developed a protein-oriented Designed high-level programming language.
Paper address: https://www.biorxiv.org/content/10.1101/2022.12.21.521526v1
Alex Rives, one of the main leaders of the research and the corresponding author of the paper "A high-level programming language for generative protein design", said on social media that this result makes the system complex and modular. Programming the generation of structures of large proteins and complexes becomes possible.
Brian Hie, one of the authors of the paper and a researcher at Stanford University, also explained the main research ideas and results of this article on Twitter. .
Overall, this article describes how generative machine learning enables the modular design of complex proteins controlled by high-level programming languages for protein design. .
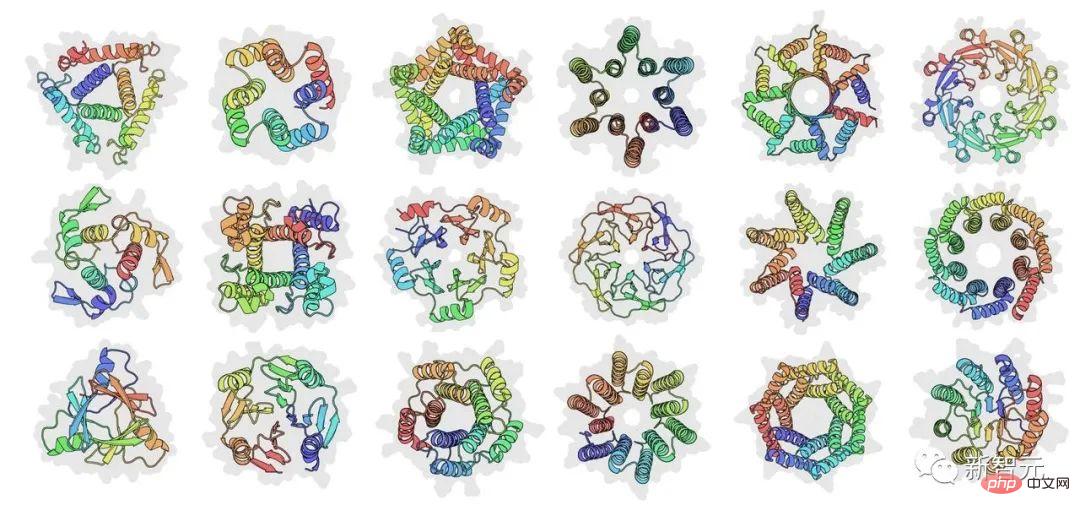
He stated that the main idea of the article is not to use building blocks of sequences or structures, but to place modularity at a higher level of abstraction and Let black box optimization generate specific designs. Atomic-level structure is predicted at every step of the optimization.
Compared with previous protein design methods, this new idea generates a method that allows designers to specify arbitrary , non-differentiable constraints, ranging from specifying atomic-level coordinates to abstract design solutions for proteins, such as symmetry designs.
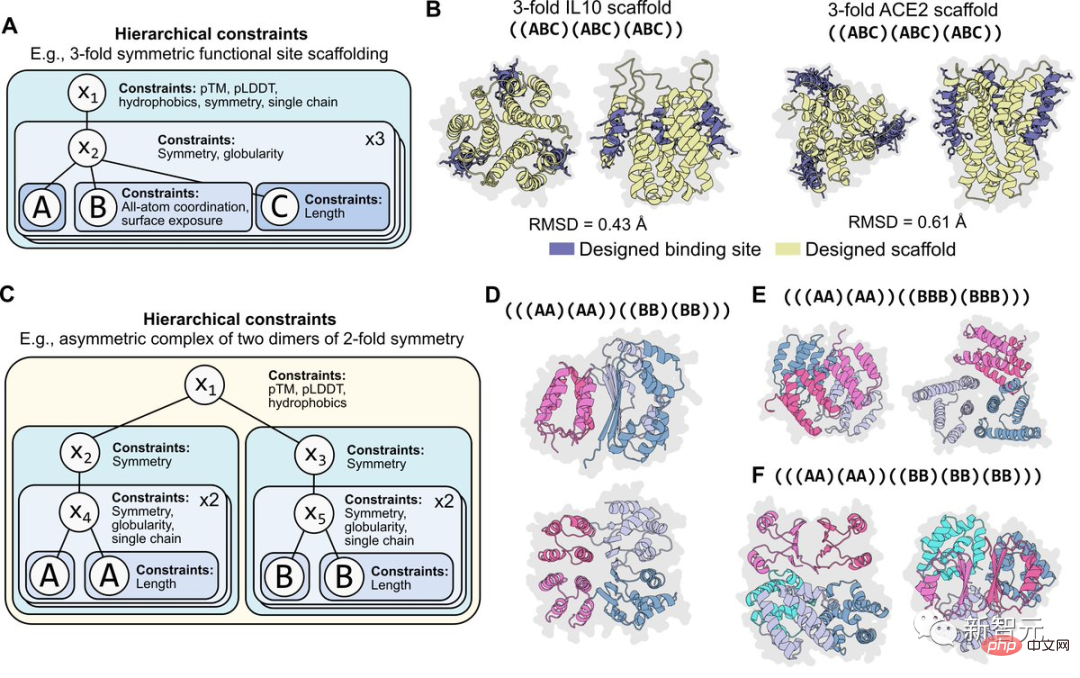
#It is important for programmability that constraints are modular. For example, the figure below shows the situation where the same constraint is applied hierarchically to two levels of symmetry programming.
#These constraints are also easy to recombine. For example, constraints on atomic coordinates can be combined with constraints on symmetry. Or different forms of two-level symmetry can be combined to program an asymmetric composite structure.
Brian Hie believes that this result is towards a more controllable, regular and expressive A step forward in protein design. He also thanked Meta AI and other collaborators for their joint efforts.
Make protein design "like building a building"
In the paper, the researchers argue that protein design would benefit from a basic set of Regularity, simplicity, and programmability are provided by abstract concepts like those used in the engineering of buildings, machines, circuits, and computer software.
#But unlike these artificial creations, proteins cannot be broken down into easily recombined parts because the local structure of the sequence is entangled with its overall environment. Together. Classic ab initio protein design attempts to identify a set of basic structural building blocks and then assemble them into higher-order structures.
#Similarly, traditional protein engineering often recombines fragments or domains of native protein sequences into hybrid chimeras. However, existing approaches are not yet able to achieve the high combinatorial complexity required for true programmability.
This paper demonstrates that modern generative models achieve the classic goals of modularity and programmability at new levels of combinatorial complexity. Putting modularity and programmability at a higher level of abstraction, generative models bridge the gap between human intuition and the generation of specific sequences and structures.
In this case, the protein designer only needs to reassemble the high-level instructions, and the task of obtaining a protein that satisfies these instructions is placed on the generative model superior.
# Researchers propose a programming language for generative protein design that allows designers to specify intuitive, modular, and hierarchical procedures. High-level programs can be transformed into low-level sequences and structures through generative models. This approach leverages advances in protein language models, which can learn structural information and design principles for proteins.
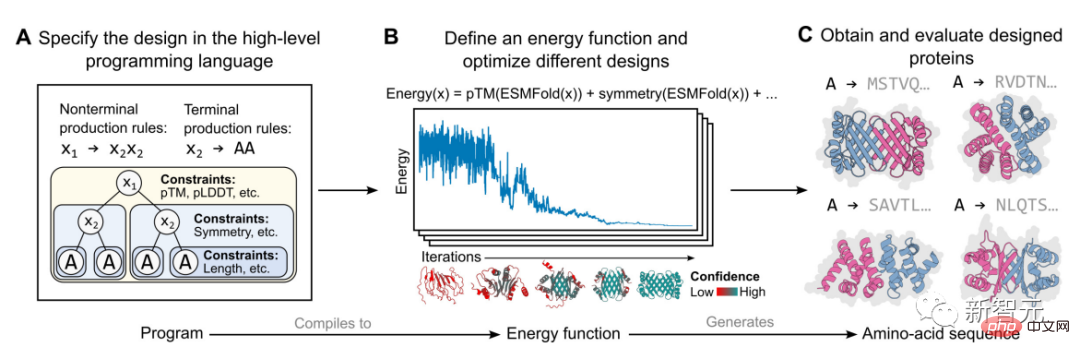
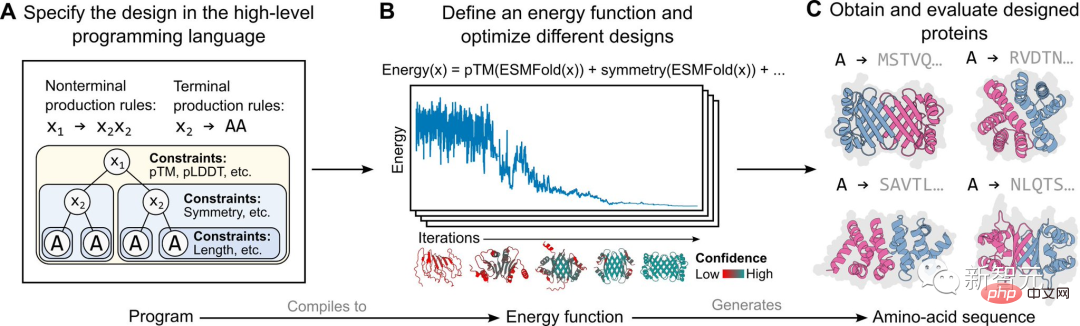
The specific implementation in this study is based on an energy-based generative model, as shown in the figure above.
First, a protein designer specifies a high-level program consisting of a set of hierarchically organized constraints (Figure A).
The program then compiles into an energy function that evaluates compatibility with constraints, which can be arbitrary and indistinguishable (Figure B ).
#Apply structural constraints by incorporating atomic-level structure predictions (enabled by language models) into the energy function. This approach is capable of generating a wide range of complex designs (Figure C).
Generating protein sequences from scratch
In the paper "Language models generalize beyond natural proteins", Tom Sercu, the author from the MetaAI team, said that this The work mainly accomplished two tasks.

Paper address: https://www.biorxiv.org/content/10.1101/2022.12.21.521521v1
The first item is to design the sequence for the given main chain structure. Using a language model, a successful design for all goals can be obtained with a success rate of 19/20, while a sequence design without the participation of a language model has a success rate of only 1/20.
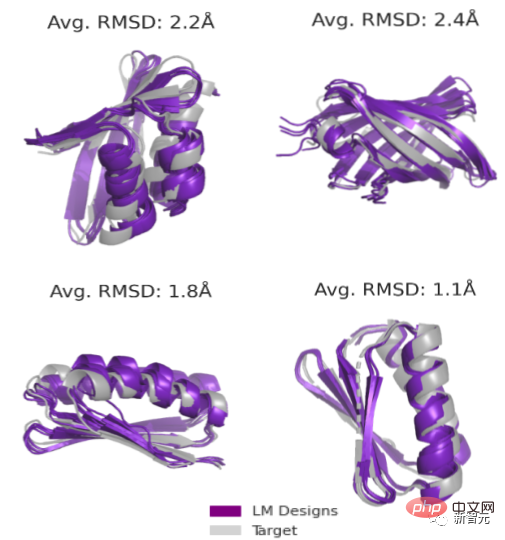
The second task is unconstrained generation. The research team proposes a new method for sampling (sequence, structure) pairs from an energy landscape defined by a language model.
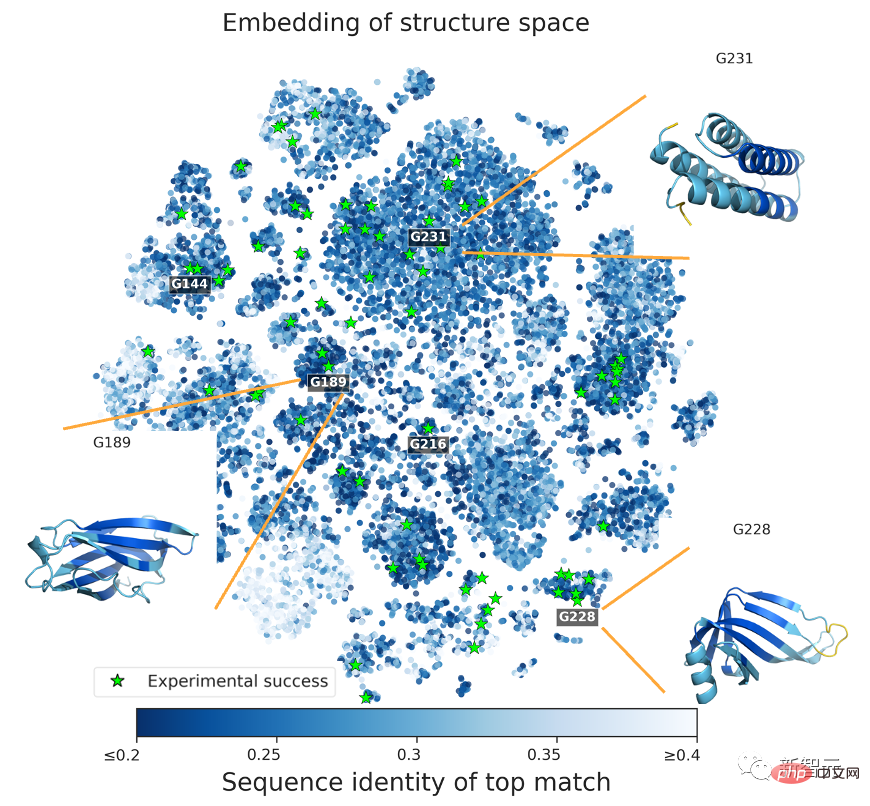
Sampling through different topologies again increases the success rate of the experiment (up to 71/129 or 55%) .
To prove that the predicted protein structure exceeds the limitations of natural proteins, the research team compared the language model generated in a sequence database covering all known natural proteins. Protein sequence search.
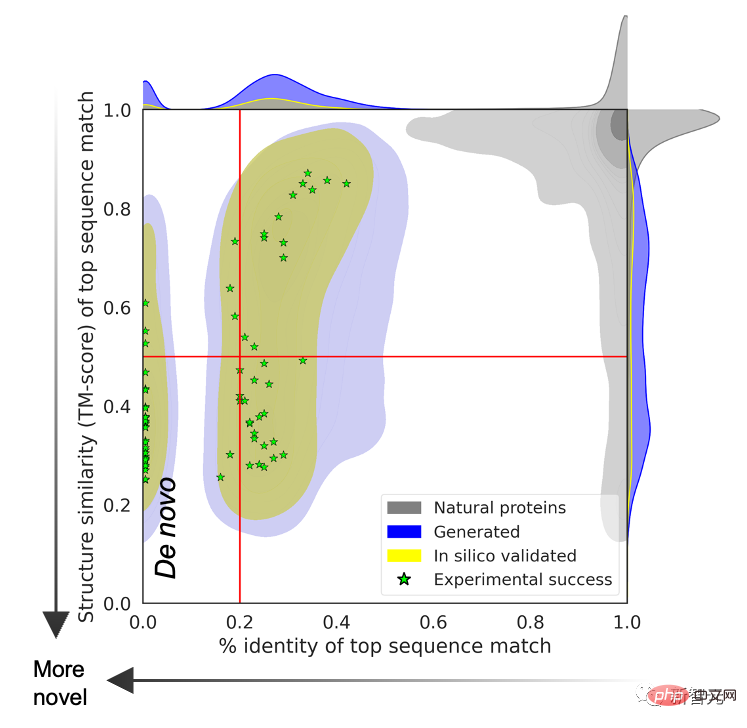
The results show that there is no matching relationship between the two, and the prediction structures generated by natural sequences and language models are different.
Sercu said that protein structures can be designed using the ESM2 protein language model alone. The research team tested 228 proteins experimentally, with a success rate of 67%!
Sercu believes that protein language models trained only on sequences can learn deep patterns connecting sequence and structure, and Can be used to design proteins de novo, beyond the design space naturally explored.
Exploring the deep grammar of protein generation
In the paper, Meta researchers stated that although the language model is only trained on sequences, The model can still design the deep grammatical structure of proteins and break through the limitations of natural proteins.
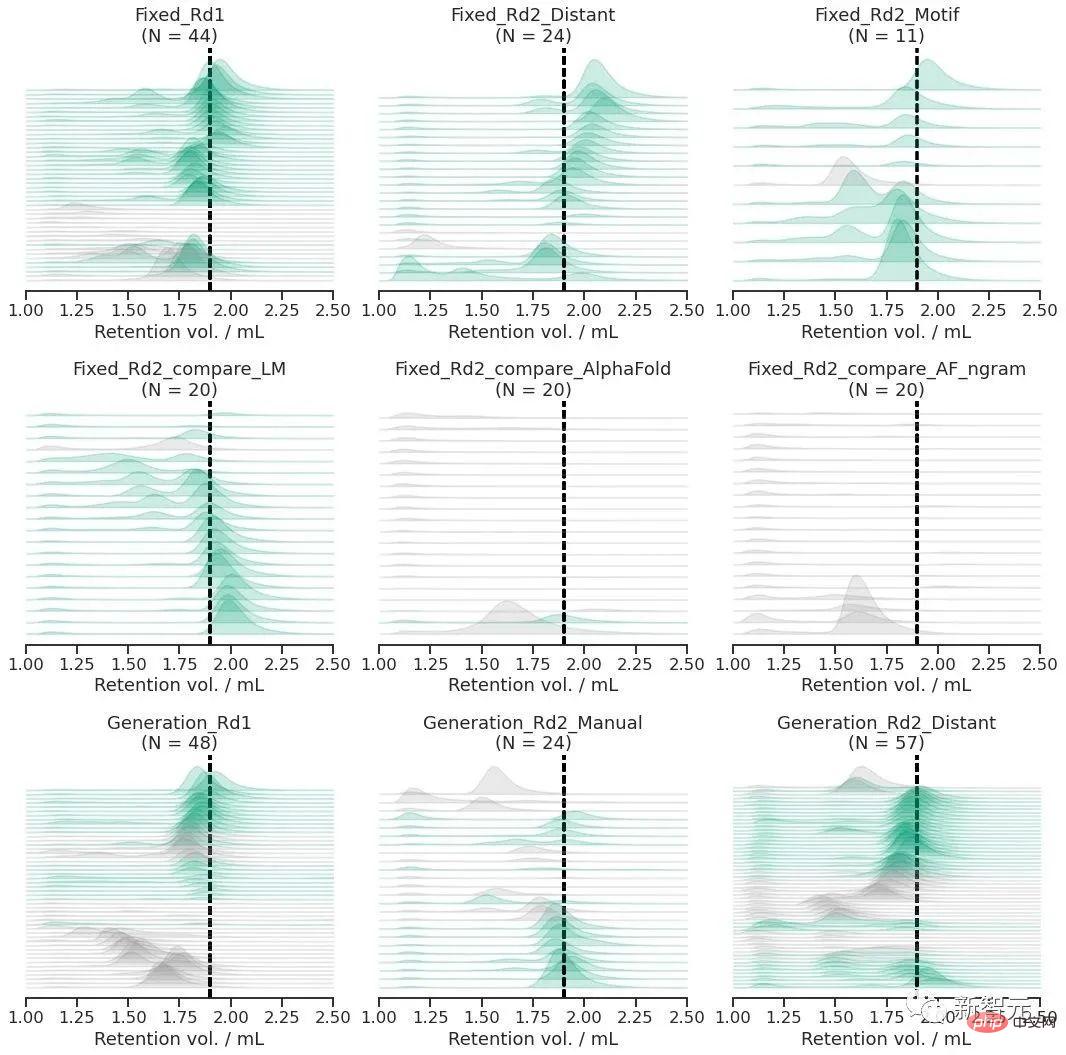
If the squares in Figure A represent the space composed of all protein sequences, then the natural protein sequence is the gray part, covering a small part of it. In order to generalize beyond natural sequences, language models need access to underlying design patterns.
What the research team has to do is two things: first, design the protein (de novo) main chain from scratch; second, based on the main chain, start from scratch to generate protein sequences.
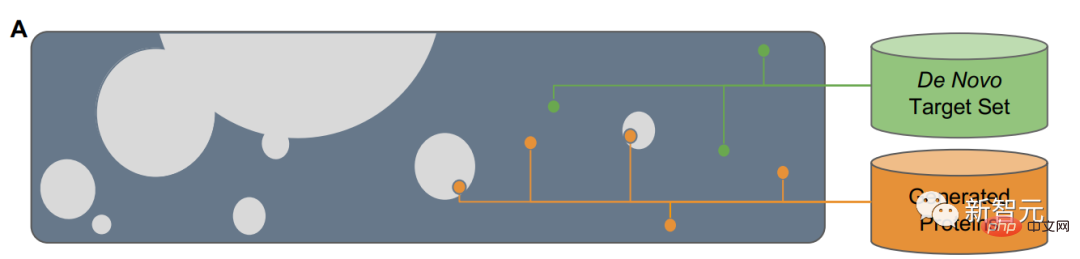
The research team used a masked language model to train ESM2, and the training content included millions of different natural proteins during the evolution process.
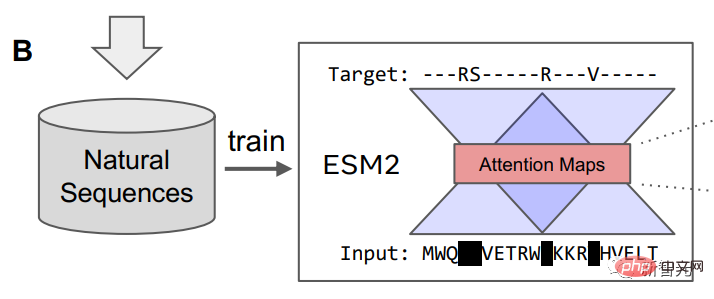
After the language model is trained, information about the tertiary structure of the protein can be identified in the internal attention state of the model. Afterwards, the researchers converted the attention of a pair of positions in the protein sequence into a distribution of distances between residues through linear projection.
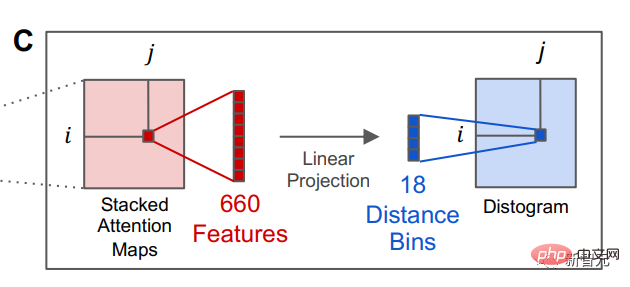
The ability of language models to predict protein structures points to the deeper structures underlying natural protein sequences, the researchers said. sequence, and the possibility that there is a deep grammar that can be learned by the model.
The results show that during the evolution process, a large number of protein sequences contain biological structures and functions, revealing the design structure of proteins. This construction is entirely reproducible by learning machine models of protein sequences.
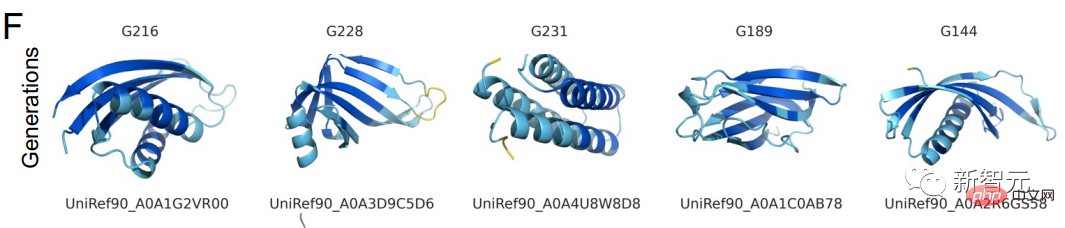
Protein structure successfully predicted by the language model in 6 experiments
The existence of deep grammars across proteins explains two seemingly contradictory sets of findings: that understanding native proteins depends on training data; and that language models can operate outside known native protein families. Predict and explore.
#If the scaling law of protein language models continues to be effective, it can be expected that the generation capabilities of AI language models will continue to improve.
The research team stated that due to the existence of the basic grammar of protein structure, the machine model will learn more rare protein structures, thereby expanding the model's prediction ability and exploration space.
# One year ago, DeepMind’s open source AlphaFold2 was launched in Nature and Science, overwhelming the biological and AI academic circles.
# One year later, artificial intelligence prediction models have sprung up, frequently filling gaps in the field of protein structure.
If humans give life to artificial intelligence, then is artificial intelligence the last piece of the puzzle for humans to complete the mystery of life?
The above is the detailed content of Meta lets a 15 billion parameter language model learn to design 'new' proteins from scratch! LeCun: Amazing results. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 The world's most powerful open source MoE model is here, with Chinese capabilities comparable to GPT-4, and the price is only nearly one percent of GPT-4-Turbo
May 07, 2024 pm 04:13 PM
The world's most powerful open source MoE model is here, with Chinese capabilities comparable to GPT-4, and the price is only nearly one percent of GPT-4-Turbo
May 07, 2024 pm 04:13 PM
Imagine an artificial intelligence model that not only has the ability to surpass traditional computing, but also achieves more efficient performance at a lower cost. This is not science fiction, DeepSeek-V2[1], the world’s most powerful open source MoE model is here. DeepSeek-V2 is a powerful mixture of experts (MoE) language model with the characteristics of economical training and efficient inference. It consists of 236B parameters, 21B of which are used to activate each marker. Compared with DeepSeek67B, DeepSeek-V2 has stronger performance, while saving 42.5% of training costs, reducing KV cache by 93.3%, and increasing the maximum generation throughput to 5.76 times. DeepSeek is a company exploring general artificial intelligence
 Hello, electric Atlas! Boston Dynamics robot comes back to life, 180-degree weird moves scare Musk
Apr 18, 2024 pm 07:58 PM
Hello, electric Atlas! Boston Dynamics robot comes back to life, 180-degree weird moves scare Musk
Apr 18, 2024 pm 07:58 PM
Boston Dynamics Atlas officially enters the era of electric robots! Yesterday, the hydraulic Atlas just "tearfully" withdrew from the stage of history. Today, Boston Dynamics announced that the electric Atlas is on the job. It seems that in the field of commercial humanoid robots, Boston Dynamics is determined to compete with Tesla. After the new video was released, it had already been viewed by more than one million people in just ten hours. The old people leave and new roles appear. This is a historical necessity. There is no doubt that this year is the explosive year of humanoid robots. Netizens commented: The advancement of robots has made this year's opening ceremony look like a human, and the degree of freedom is far greater than that of humans. But is this really not a horror movie? At the beginning of the video, Atlas is lying calmly on the ground, seemingly on his back. What follows is jaw-dropping
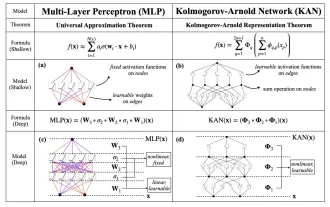 KAN, which replaces MLP, has been extended to convolution by open source projects
Jun 01, 2024 pm 10:03 PM
KAN, which replaces MLP, has been extended to convolution by open source projects
Jun 01, 2024 pm 10:03 PM
Earlier this month, researchers from MIT and other institutions proposed a very promising alternative to MLP - KAN. KAN outperforms MLP in terms of accuracy and interpretability. And it can outperform MLP running with a larger number of parameters with a very small number of parameters. For example, the authors stated that they used KAN to reproduce DeepMind's results with a smaller network and a higher degree of automation. Specifically, DeepMind's MLP has about 300,000 parameters, while KAN only has about 200 parameters. KAN has a strong mathematical foundation like MLP. MLP is based on the universal approximation theorem, while KAN is based on the Kolmogorov-Arnold representation theorem. As shown in the figure below, KAN has
 Tesla robots work in factories, Musk: The degree of freedom of hands will reach 22 this year!
May 06, 2024 pm 04:13 PM
Tesla robots work in factories, Musk: The degree of freedom of hands will reach 22 this year!
May 06, 2024 pm 04:13 PM
The latest video of Tesla's robot Optimus is released, and it can already work in the factory. At normal speed, it sorts batteries (Tesla's 4680 batteries) like this: The official also released what it looks like at 20x speed - on a small "workstation", picking and picking and picking: This time it is released One of the highlights of the video is that Optimus completes this work in the factory, completely autonomously, without human intervention throughout the process. And from the perspective of Optimus, it can also pick up and place the crooked battery, focusing on automatic error correction: Regarding Optimus's hand, NVIDIA scientist Jim Fan gave a high evaluation: Optimus's hand is the world's five-fingered robot. One of the most dexterous. Its hands are not only tactile
 FisheyeDetNet: the first target detection algorithm based on fisheye camera
Apr 26, 2024 am 11:37 AM
FisheyeDetNet: the first target detection algorithm based on fisheye camera
Apr 26, 2024 am 11:37 AM
Target detection is a relatively mature problem in autonomous driving systems, among which pedestrian detection is one of the earliest algorithms to be deployed. Very comprehensive research has been carried out in most papers. However, distance perception using fisheye cameras for surround view is relatively less studied. Due to large radial distortion, standard bounding box representation is difficult to implement in fisheye cameras. To alleviate the above description, we explore extended bounding box, ellipse, and general polygon designs into polar/angular representations and define an instance segmentation mIOU metric to analyze these representations. The proposed model fisheyeDetNet with polygonal shape outperforms other models and simultaneously achieves 49.5% mAP on the Valeo fisheye camera dataset for autonomous driving
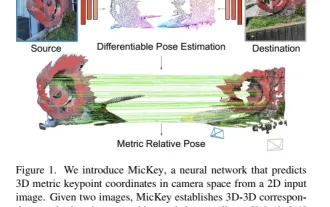 The latest from Oxford University! Mickey: 2D image matching in 3D SOTA! (CVPR\'24)
Apr 23, 2024 pm 01:20 PM
The latest from Oxford University! Mickey: 2D image matching in 3D SOTA! (CVPR\'24)
Apr 23, 2024 pm 01:20 PM
Project link written in front: https://nianticlabs.github.io/mickey/ Given two pictures, the camera pose between them can be estimated by establishing the correspondence between the pictures. Typically, these correspondences are 2D to 2D, and our estimated poses are scale-indeterminate. Some applications, such as instant augmented reality anytime, anywhere, require pose estimation of scale metrics, so they rely on external depth estimators to recover scale. This paper proposes MicKey, a keypoint matching process capable of predicting metric correspondences in 3D camera space. By learning 3D coordinate matching across images, we are able to infer metric relative
 New affordable Meta Quest 3S VR headset appears on FCC, suggesting imminent launch
Sep 04, 2024 am 06:51 AM
New affordable Meta Quest 3S VR headset appears on FCC, suggesting imminent launch
Sep 04, 2024 am 06:51 AM
The Meta Connect 2024event is set for September 25 to 26, and in this event, the company is expected to unveil a new affordable virtual reality headset. Rumored to be the Meta Quest 3S, the VR headset has seemingly appeared on FCC listing. This sugge
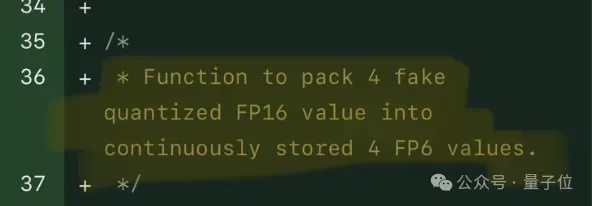 Single card running Llama 70B is faster than dual card, Microsoft forced FP6 into A100 | Open source
Apr 29, 2024 pm 04:55 PM
Single card running Llama 70B is faster than dual card, Microsoft forced FP6 into A100 | Open source
Apr 29, 2024 pm 04:55 PM
FP8 and lower floating point quantification precision are no longer the "patent" of H100! Lao Huang wanted everyone to use INT8/INT4, and the Microsoft DeepSpeed team started running FP6 on A100 without official support from NVIDIA. Test results show that the new method TC-FPx's FP6 quantization on A100 is close to or occasionally faster than INT4, and has higher accuracy than the latter. On top of this, there is also end-to-end large model support, which has been open sourced and integrated into deep learning inference frameworks such as DeepSpeed. This result also has an immediate effect on accelerating large models - under this framework, using a single card to run Llama, the throughput is 2.65 times higher than that of dual cards. one





