 Technology peripherals
Technology peripherals
 AI
AI
 Introducing ImageMol, the world's first molecular image generation framework based on self-supervised learning
Introducing ImageMol, the world's first molecular image generation framework based on self-supervised learning
Introducing ImageMol, the world's first molecular image generation framework based on self-supervised learning
Molecular is the smallest unit that maintains the chemical stability of a substance. The study of molecules is a fundamental issue in many scientific fields such as pharmacy, materials science, biology, and chemistry.
Molecular Representation Learning has been a very popular direction in recent years and can currently be divided into many schools:
- Computational pharmacologists say: Molecules can be represented as a string of fingerprints, or descriptors, such as AttentiveFP proposed by Shanghai Pharmaceuticals, which is an outstanding representative in this regard.
- NLPer said: Molecules can be expressed as SMILES (sequences) and then processed as natural language, such as Baidu's X-Mol, which is an outstanding representative in this regard.
- Graph neural network researchers say: Molecules can be represented as a graph (Graph), which is an adjacency matrix, and then processed using graph neural networks, such as Tencent's GROVER, MIT's DMPNN, Methods such as CMU's MOLCLR are outstanding representatives in this regard.
However, current characterization methods still have some limitations. For example, sequence representation lacks explicit structural information of molecules, and the expression ability of existing graph neural networks still has many limitations (Teacher Shen Huawei from the Institute of Computing Technology, Chinese Academy of Sciences discussed this, see Mr. Shen’s report "The Expression Ability of Graph Neural Networks").
What’s interesting is that when we study molecules in high school chemistry, we see images of molecules. When chemists design molecules, they also observe and think based on molecular images. A natural idea arises spontaneously: "Why not directly use molecular images to represent molecules?"If images can be used directly to represent molecules, then in CV (Computer Vision) Can't all the eighteen martial arts be used to study molecules?
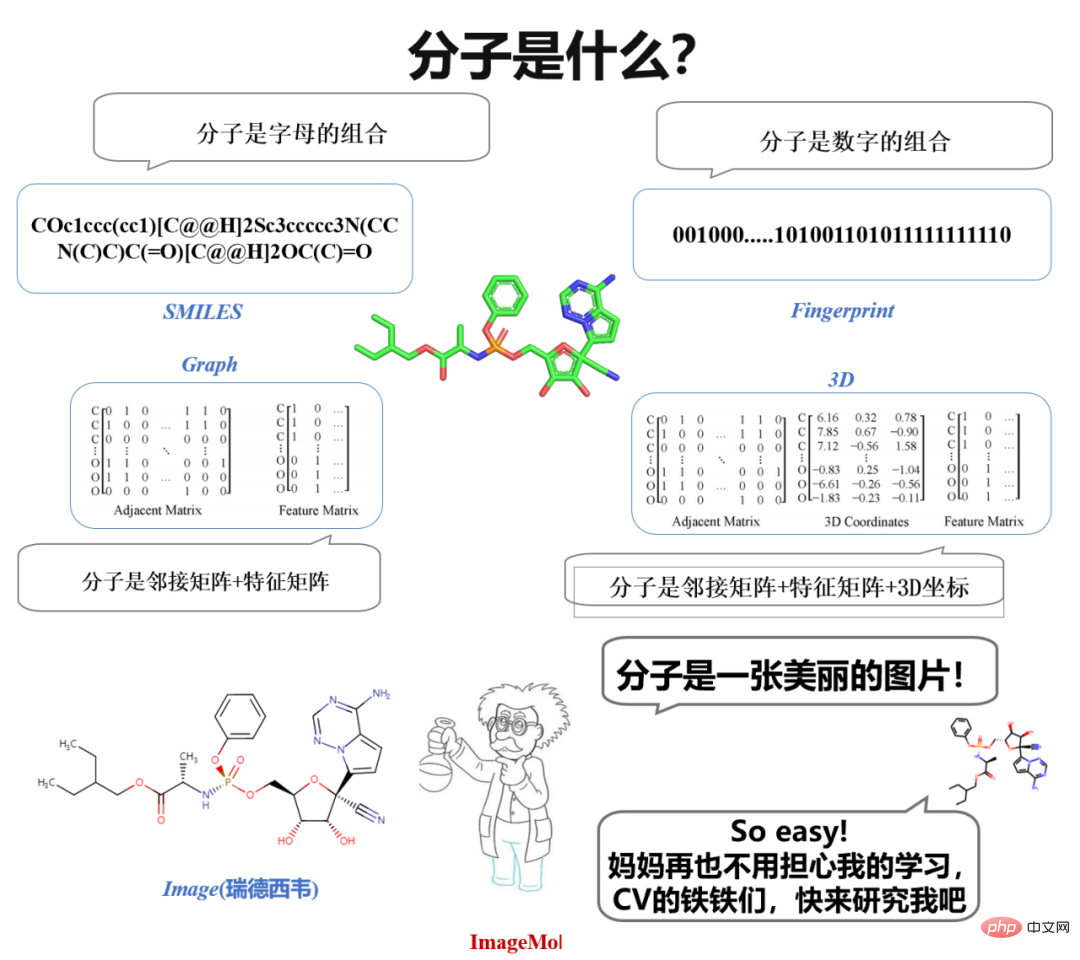
Just do it. There are so many models in CV, why don’t you use them to learn molecules? Stop, there is another important issue - data! Especially labeled data! In the field of CV, data annotation does not seem to be difficult. For classic CV and NLP problems such as image recognition or emotion classification, a person can annotate an average of 800 pieces of data. However, in the molecular field, molecular properties can only be assessed through wet experiments and clinical experiments, so labeled data are very scarce.
Based on this, researchers from Hunan University proposed the world's first unsupervised learning framework for molecular images, ImageMol, which uses large-scale unlabeled molecular image data for unsupervised pre-training. It provides a new paradigm for understanding molecular properties and drug targets, proving that molecular images have great potential in the field of intelligent drug research and development. The result was published in the top international journal "Nature Machine Intelligence" under the title "Accurate prediction of molecular properties and drug targets using a self-supervised image representation learning framework". The success achieved at the intersection of computer vision and molecular fields demonstrates the great potential of using computer vision technology to understand molecular properties and drug target mechanisms, and provides new opportunities for research in the molecular field.
Paper link: https://www.nature.com/articles/s42256-022-00557-6.pdf
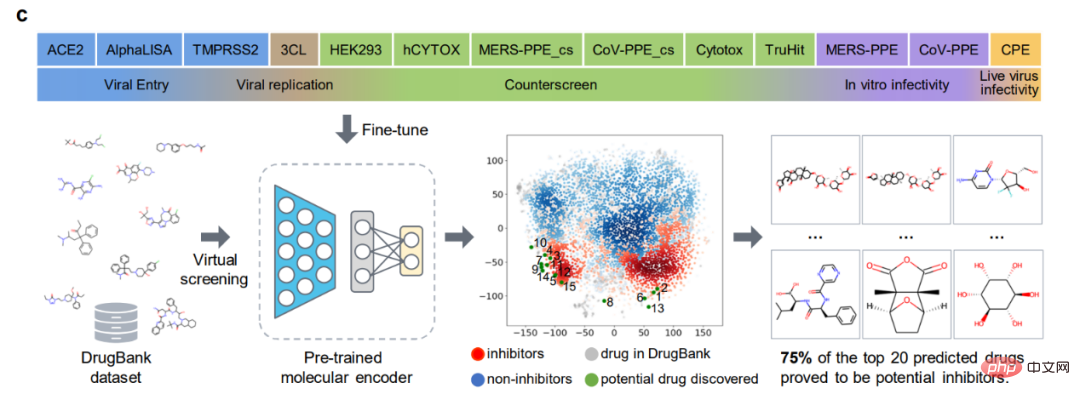
ImageMol model structure
The overall structure of ImageMol is shown in the figure below, which is divided into three parts:
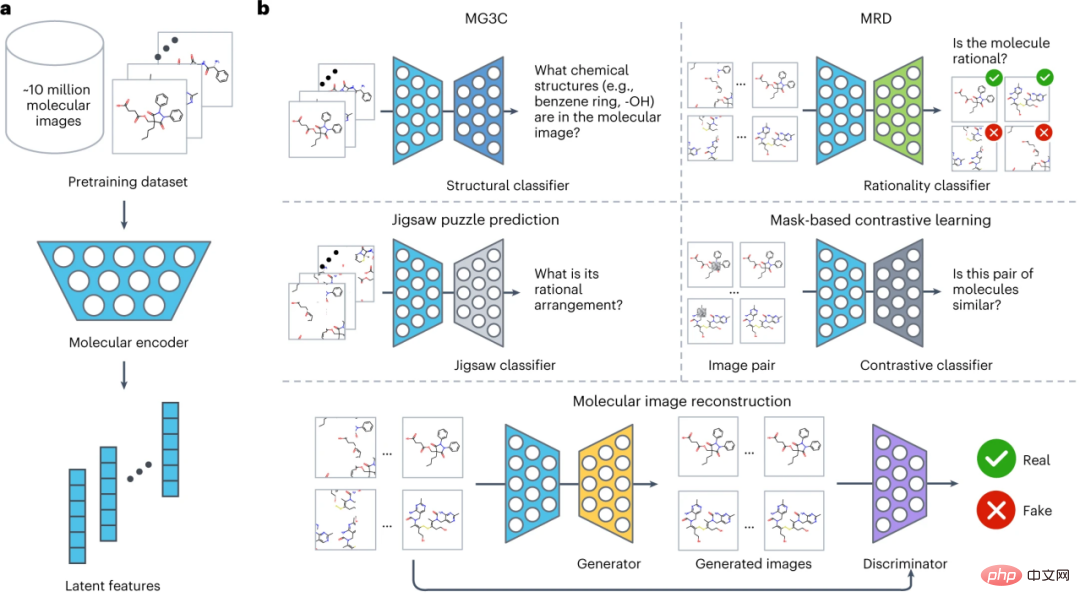
(1) Design a molecular encoder ResNet18 (light blue), which can extract latent features from about 10 million molecular images (a).
(2) Considering the chemical knowledge and structural information in the molecular image, five pre-training strategies (MG3C, MRD, JPP, MCL, MIR) are used to optimize the latent representation of the molecular encoder (b). Specifically:
① MG3C (Muti-granularity chemical clusters classification): The structure classifier (dark blue) is used to predict molecular images Chemical structure information;
② MRD (Molecular rationality discrimination): the rationality classifier (green), which is used to distinguish between reasonable and unreasonable molecules;
③ JPP (Jigsaw puzzle prediction): The Jigsaw classifier (light gray) is used to predict the reasonable arrangement of molecules;
④ MCL (MASK-based contrastive learning MASK-based contrastive learning): The contrastive classifier (dark gray) is used to maximize the similarity between the original image and the mask image;
⑤ MIR (Molecular image reconstruction): The generator (yellow) is used to restore latent features to the molecular image, and the discriminator (purple) is used to distinguish between real images and generated images. Fake molecular images generated by the machine.
(3) Fine-tune the preprocessed molecular encoder in downstream tasks to further improve model performance (c).
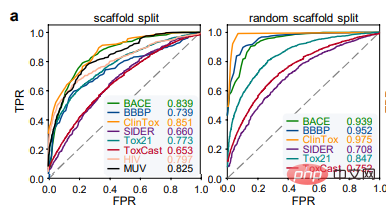
Benchmark Evaluation
The authors first evaluated the performance of ImageMol using 8 drug discovery benchmark datasets and used two The most popular splitting strategies (scaffold split and random scaffold split) are used to evaluate the performance of ImageMol on all benchmark datasets. In the classification task, the Receiver Operating Characteristic (ROC) curve and the Area Under Curve (AUC) are used to evaluate. From the experimental results, it can be seen that ImageMol can obtain higher AUC values. (Figure a).
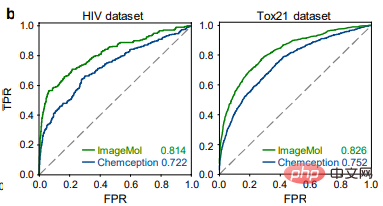
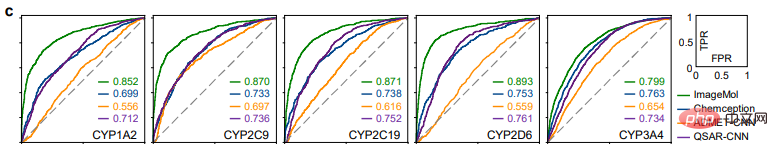
Comparison of the detection results of HIV and Tox21 between ImageMol and Chemception, a classic convolutional neural network framework for predicting molecular images (Figure b), ImageMol’s AUC Value is higher. This article further evaluates the performance of ImageMol in predicting drug metabolism by five major metabolizing enzymes: CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4. Figure c shows that ImageMol achieves better results compared with three state-of-the-art molecular image-based representation models (Chemception46, ADMET-CNN12 and QSAR-CNN47) in the prediction of inhibitors versus non-inhibitors of five major drug metabolizing enzymes. achieved higher AUC values (ranging from 0.799 to 0.893).
This paper further compares the performance of ImageMol with three state-of-the-art molecular representation models, e.g. As shown in Figures d and e. ImageMol has better performance compared to fingerprint-based models (such as AttentiveFP), sequence-based models (such as TF_Robust), and graph-based models (such as N-GRAM, GROVER, and MPG) that use random skeleton partitioning. Furthermore, ImageMol achieved higher AUC values on CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4 compared with traditional MACCS-based methods and FP4-based methods (Figure f).
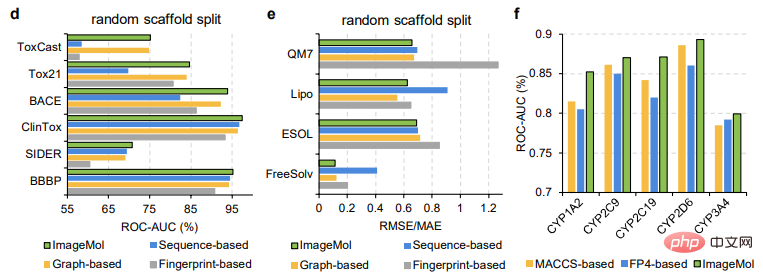
ImageMol is compared with sequence-based models (including RNN_LR, TRFM_LR, RNN_MLP, TRFM_MLP, RNN_RF, TRFM_RF, and CHEM-BERT) and graph-based models (including MolCLRGIN, MolCLRGCN, and GROVER), as shown in Figure g It shows that ImageMol achieves better AUC performance on CYP1A2, CYP2C9, CYP2C19, CYP2D6, and CYP3A4.
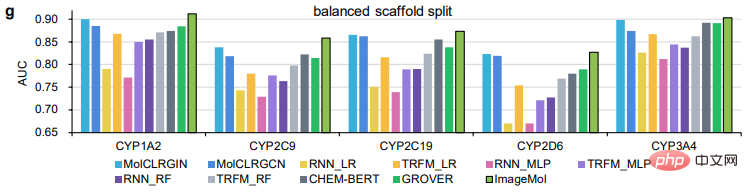
In the above comparison between ImageMol and other advanced models, we can see the superiority of ImageMol.
Since the outbreak of COVID-19, we have urgently needed to develop effective treatment strategies for COVID-19. Therefore, the authors evaluated ImageMol accordingly in this aspect.
Prediction of 13 SARS-CoV-2 targets
ImageMol conducted prediction experiments on 13 SARS-CoV-2 targets that are of concern today. -CoV-2 bioassay data set, ImageMol achieved high AUC values of 72.6% to 83.7%. Panel a reveals the potential signature identified by ImageMol, which clusters well on 13 targets or endpoints active and inactive anti-SARS-CoV-2, with higher AUC values than the other The model Jure's GNN is more than 12% higher, reflecting the high accuracy and strong generalization of the model.
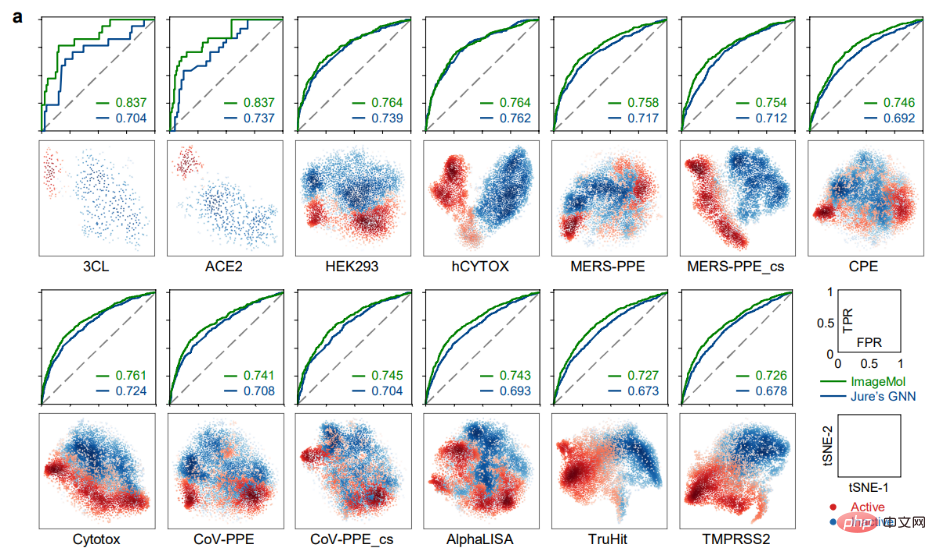
Identification of anti-SARS-CoV-2 inhibitors
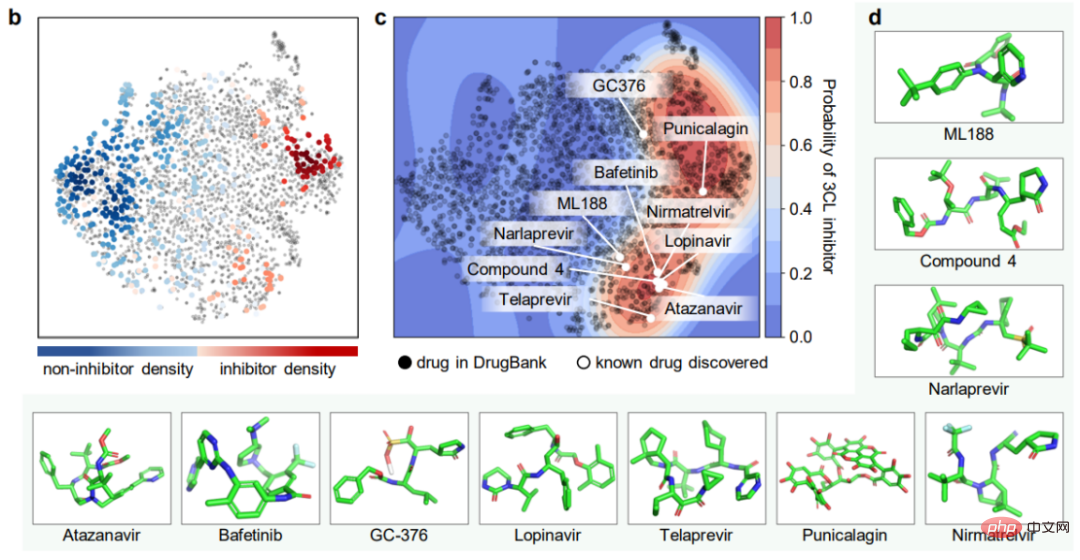
The most direct experiment related to the study of drug molecules is here, using ImageMol Directly identify inhibitor molecules! Through the molecular image representation of inhibitors and non-inhibitors of 3CL protease (which has been proven to be a promising therapeutic development target for the treatment of COVID-19) under the ImageMol framework, this study found that 3CL inhibitors and non-inhibitors have significant differences in t- Well separated in the SNE plot, as shown in Figure b below.
In addition, ImageMol identified 10 of the 16 known 3CL protease inhibitors and visualized these 10 drugs into the embedded space in the figure (success rate 62.5%) , indicating high generalization ability in anti-SARS-CoV-2 drug discovery. When using the HEY293 assay to predict anti-SARS-CoV-2 repurposed drugs, ImageMol successfully predicted 42 out of 70 drugs (60% success rate), indicating that ImageMol is also good at inferring potential drug candidates in the HEY293 assay. It has high promotion potential. Figure c below shows ImageMol’s discovery of drugs that are potential inhibitors of 3CL on the DrugBank dataset. Panel d shows the molecular structure of the 3CL inhibitor discovered by ImageMol.
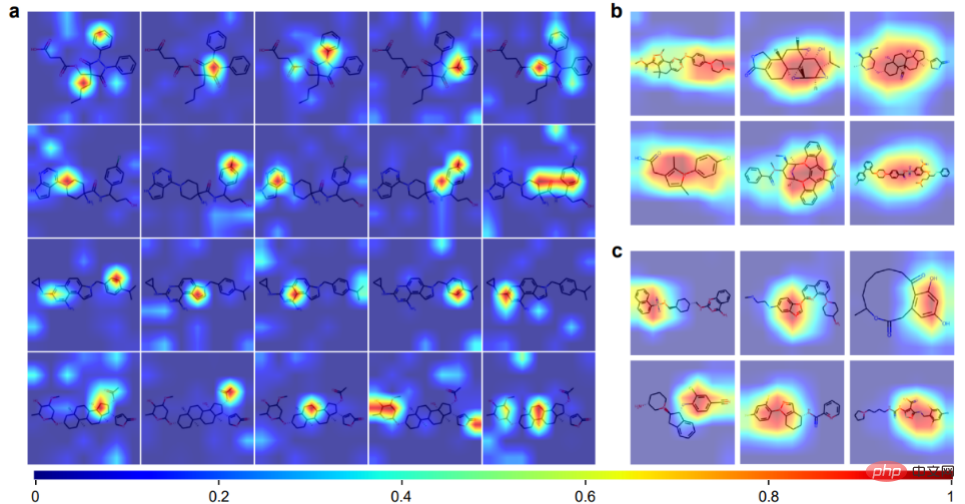
Attention Visualization
ImageMol can obtain prior knowledge of chemical information from molecular image representations, including = O bonds, -OH bond, -NH3 bond and benzene ring. Panels b and c show 12 example molecules visualized by ImageMol's Grad-CAM. This means that ImageMol accurately captures attention to both global (b) and local (c) structural information simultaneously. These results allow researchers to visually understand how molecular structure affects properties and targets.
The above is the detailed content of Introducing ImageMol, the world's first molecular image generation framework based on self-supervised learning. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1664
1664
 14
14
 1423
1423
 52
52
 1317
1317
 25
25
 1268
1268
 29
29
 1242
1242
 24
24
 The world's most powerful open source MoE model is here, with Chinese capabilities comparable to GPT-4, and the price is only nearly one percent of GPT-4-Turbo
May 07, 2024 pm 04:13 PM
The world's most powerful open source MoE model is here, with Chinese capabilities comparable to GPT-4, and the price is only nearly one percent of GPT-4-Turbo
May 07, 2024 pm 04:13 PM
Imagine an artificial intelligence model that not only has the ability to surpass traditional computing, but also achieves more efficient performance at a lower cost. This is not science fiction, DeepSeek-V2[1], the world’s most powerful open source MoE model is here. DeepSeek-V2 is a powerful mixture of experts (MoE) language model with the characteristics of economical training and efficient inference. It consists of 236B parameters, 21B of which are used to activate each marker. Compared with DeepSeek67B, DeepSeek-V2 has stronger performance, while saving 42.5% of training costs, reducing KV cache by 93.3%, and increasing the maximum generation throughput to 5.76 times. DeepSeek is a company exploring general artificial intelligence
 AI subverts mathematical research! Fields Medal winner and Chinese-American mathematician led 11 top-ranked papers | Liked by Terence Tao
Apr 09, 2024 am 11:52 AM
AI subverts mathematical research! Fields Medal winner and Chinese-American mathematician led 11 top-ranked papers | Liked by Terence Tao
Apr 09, 2024 am 11:52 AM
AI is indeed changing mathematics. Recently, Tao Zhexuan, who has been paying close attention to this issue, forwarded the latest issue of "Bulletin of the American Mathematical Society" (Bulletin of the American Mathematical Society). Focusing on the topic "Will machines change mathematics?", many mathematicians expressed their opinions. The whole process was full of sparks, hardcore and exciting. The author has a strong lineup, including Fields Medal winner Akshay Venkatesh, Chinese mathematician Zheng Lejun, NYU computer scientist Ernest Davis and many other well-known scholars in the industry. The world of AI has changed dramatically. You know, many of these articles were submitted a year ago.
 Google is ecstatic: JAX performance surpasses Pytorch and TensorFlow! It may become the fastest choice for GPU inference training
Apr 01, 2024 pm 07:46 PM
Google is ecstatic: JAX performance surpasses Pytorch and TensorFlow! It may become the fastest choice for GPU inference training
Apr 01, 2024 pm 07:46 PM
The performance of JAX, promoted by Google, has surpassed that of Pytorch and TensorFlow in recent benchmark tests, ranking first in 7 indicators. And the test was not done on the TPU with the best JAX performance. Although among developers, Pytorch is still more popular than Tensorflow. But in the future, perhaps more large models will be trained and run based on the JAX platform. Models Recently, the Keras team benchmarked three backends (TensorFlow, JAX, PyTorch) with the native PyTorch implementation and Keras2 with TensorFlow. First, they select a set of mainstream
 Hello, electric Atlas! Boston Dynamics robot comes back to life, 180-degree weird moves scare Musk
Apr 18, 2024 pm 07:58 PM
Hello, electric Atlas! Boston Dynamics robot comes back to life, 180-degree weird moves scare Musk
Apr 18, 2024 pm 07:58 PM
Boston Dynamics Atlas officially enters the era of electric robots! Yesterday, the hydraulic Atlas just "tearfully" withdrew from the stage of history. Today, Boston Dynamics announced that the electric Atlas is on the job. It seems that in the field of commercial humanoid robots, Boston Dynamics is determined to compete with Tesla. After the new video was released, it had already been viewed by more than one million people in just ten hours. The old people leave and new roles appear. This is a historical necessity. There is no doubt that this year is the explosive year of humanoid robots. Netizens commented: The advancement of robots has made this year's opening ceremony look like a human, and the degree of freedom is far greater than that of humans. But is this really not a horror movie? At the beginning of the video, Atlas is lying calmly on the ground, seemingly on his back. What follows is jaw-dropping
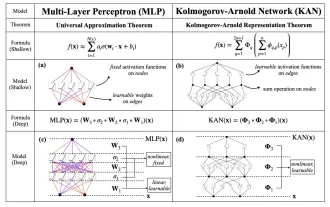 KAN, which replaces MLP, has been extended to convolution by open source projects
Jun 01, 2024 pm 10:03 PM
KAN, which replaces MLP, has been extended to convolution by open source projects
Jun 01, 2024 pm 10:03 PM
Earlier this month, researchers from MIT and other institutions proposed a very promising alternative to MLP - KAN. KAN outperforms MLP in terms of accuracy and interpretability. And it can outperform MLP running with a larger number of parameters with a very small number of parameters. For example, the authors stated that they used KAN to reproduce DeepMind's results with a smaller network and a higher degree of automation. Specifically, DeepMind's MLP has about 300,000 parameters, while KAN only has about 200 parameters. KAN has a strong mathematical foundation like MLP. MLP is based on the universal approximation theorem, while KAN is based on the Kolmogorov-Arnold representation theorem. As shown in the figure below, KAN has
 Tesla robots work in factories, Musk: The degree of freedom of hands will reach 22 this year!
May 06, 2024 pm 04:13 PM
Tesla robots work in factories, Musk: The degree of freedom of hands will reach 22 this year!
May 06, 2024 pm 04:13 PM
The latest video of Tesla's robot Optimus is released, and it can already work in the factory. At normal speed, it sorts batteries (Tesla's 4680 batteries) like this: The official also released what it looks like at 20x speed - on a small "workstation", picking and picking and picking: This time it is released One of the highlights of the video is that Optimus completes this work in the factory, completely autonomously, without human intervention throughout the process. And from the perspective of Optimus, it can also pick up and place the crooked battery, focusing on automatic error correction: Regarding Optimus's hand, NVIDIA scientist Jim Fan gave a high evaluation: Optimus's hand is the world's five-fingered robot. One of the most dexterous. Its hands are not only tactile
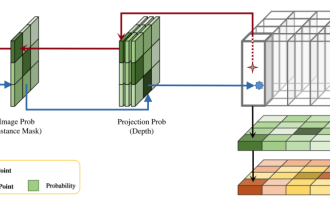 DualBEV: significantly surpassing BEVFormer and BEVDet4D, open the book!
Mar 21, 2024 pm 05:21 PM
DualBEV: significantly surpassing BEVFormer and BEVDet4D, open the book!
Mar 21, 2024 pm 05:21 PM
This paper explores the problem of accurately detecting objects from different viewing angles (such as perspective and bird's-eye view) in autonomous driving, especially how to effectively transform features from perspective (PV) to bird's-eye view (BEV) space. Transformation is implemented via the Visual Transformation (VT) module. Existing methods are broadly divided into two strategies: 2D to 3D and 3D to 2D conversion. 2D-to-3D methods improve dense 2D features by predicting depth probabilities, but the inherent uncertainty of depth predictions, especially in distant regions, may introduce inaccuracies. While 3D to 2D methods usually use 3D queries to sample 2D features and learn the attention weights of the correspondence between 3D and 2D features through a Transformer, which increases the computational and deployment time.
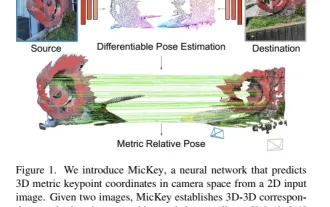 The latest from Oxford University! Mickey: 2D image matching in 3D SOTA! (CVPR\'24)
Apr 23, 2024 pm 01:20 PM
The latest from Oxford University! Mickey: 2D image matching in 3D SOTA! (CVPR\'24)
Apr 23, 2024 pm 01:20 PM
Project link written in front: https://nianticlabs.github.io/mickey/ Given two pictures, the camera pose between them can be estimated by establishing the correspondence between the pictures. Typically, these correspondences are 2D to 2D, and our estimated poses are scale-indeterminate. Some applications, such as instant augmented reality anytime, anywhere, require pose estimation of scale metrics, so they rely on external depth estimators to recover scale. This paper proposes MicKey, a keypoint matching process capable of predicting metric correspondences in 3D camera space. By learning 3D coordinate matching across images, we are able to infer metric relative



