 Technology peripherals
Technology peripherals
 AI
AI
 Comparison of common dimensionality reduction technologies: feasibility analysis of reducing data dimensions while maintaining information integrity
Comparison of common dimensionality reduction technologies: feasibility analysis of reducing data dimensions while maintaining information integrity
Comparison of common dimensionality reduction technologies: feasibility analysis of reducing data dimensions while maintaining information integrity
This article will compare the effectiveness of various dimensionality reduction techniques on tabular data in machine learning tasks. We apply dimensionality reduction methods to the dataset and evaluate their effectiveness through regression and classification analyses. We apply dimensionality reduction methods to various datasets obtained from UCI related to different domains. A total of 15 datasets were selected, 7 of which will be used for regression and 8 for classification.
To make this article easy to read and understand, only the preprocessing and analysis of one dataset is shown. The experiment starts by loading the dataset. The data set is split into training and test sets and then normalized to have a mean of 0 and a standard deviation of 1.
Dimensionality reduction techniques are then applied to the training data and the test set is transformed for dimensionality reduction using the same parameters. For regression, principal component analysis (PCA) and singular value decomposition (SVD) are used for dimensionality reduction. On the other hand, for classification, linear discriminant analysis (LDA) is used.
After dimensionality reduction, multiple machine learning models are trained Tests were conducted and the performance of different models was compared on different datasets obtained through different dimensionality reduction methods.
Let us start the process by loading the first dataset,
import pandas as pd ## for data manipulation df = pd.read_excel(r'RegressionAirQualityUCI.xlsx') print(df.shape) df.head()
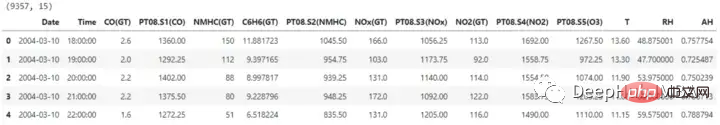
The dataset contains 15 columns, One of them is the need to predict labels. Before continuing with dimensionality reduction, the date and time columns are also removed.
X = df.drop(['CO(GT)', 'Date', 'Time'], axis=1) y = df['CO(GT)'] X.shape, y.shape #Output: ((9357, 12), (9357,))
For training, we need to divide the data set into a training set and a test set, so that the effectiveness of the dimensionality reduction method and the machine learning model trained on the dimensionality reduction feature space can be evaluated. The model will be trained using the training set and performance will be evaluated using the test set.
from sklearn.model_selection import train_test_split X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2) X_train.shape, X_test.shape, y_train.shape, y_test.shape #Output: ((7485, 12), (1872, 12), (7485,), (1872,))
Before using dimensionality reduction techniques on the data set, the input data can be scaled to ensure that all features are on the same scale. This is critical for linear models because some dimensionality reduction methods can change their output depending on whether the data is normalized and are sensitive to the size of the features.
from sklearn.preprocessing import StandardScaler scaler = StandardScaler() X_train = scaler.fit_transform(X_train) X_test = scaler.transform(X_test) X_train.shape, X_test.shape
Principal Component Analysis (PCA)
The PCA method of linear dimensionality reduction reduces the dimensionality of the data while retaining as much data variance as possible.
The PCA method of the Python sklearn.decomposition module will be used here. The number of components to retain is specified via this parameter, and this number affects how many dimensions are included in the smaller feature space. As an alternative, we can set a target variance to retain, which establishes the number of components based on the amount of variance in the captured data, which we set here to 0.95
from sklearn.decomposition import PCA pca = PCA(n_compnotallow=0.95) X_train_pca = pca.fit_transform(X_train) X_test_pca = pca.transform(X_test) X_train_pca

What do the above features represent? Principal component analysis (PCA) projects the data into a low-dimensional space, trying to retain as many differences in the data as possible. While this may help with specific operations, it may also make the data more difficult to understand. , PCA can identify new axes in the data that are linear fusions of the initial features.
Singular Value Decomposition (SVD)
SVD is a linear dimensionality reduction technique that projects features with small data variance into a low-dimensional space. We need to set the number of components to retain after dimensionality reduction. Here we will reduce the dimensionality by 2/3.
from sklearn.decomposition import TruncatedSVD svd = TruncatedSVD(n_compnotallow=int(X_train.shape[1]*0.33)) X_train_svd = svd.fit_transform(X_train) X_test_svd = svd.transform(X_test) X_train_svd

Training the regression model
Now, we will start training and testing the model using the above three types of data (original dataset, PCA and SVD) , and we use multiple models for comparison.
import numpy as np from sklearn.linear_model import LinearRegression from sklearn.neighbors import KNeighborsRegressor from sklearn.svm import SVR from sklearn.tree import DecisionTreeRegressor from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor from sklearn.metrics import r2_score, mean_squared_error import time
train_test_ML: This function will complete the repetitive tasks related to the training and testing of the model. The performance of all models was evaluated by calculating rmse and r2_score. and returns a dataset with all details and calculated values. It will also log the time each model took to train and test on its respective dataset.
def train_test_ML(dataset, dataform, X_train, y_train, X_test, y_test):
temp_df = pd.DataFrame(columns=['Data Set', 'Data Form', 'Dimensions', 'Model', 'R2 Score', 'RMSE', 'Time Taken'])
for i in [LinearRegression, KNeighborsRegressor, SVR, DecisionTreeRegressor, RandomForestRegressor, GradientBoostingRegressor]:
start_time = time.time()
reg = i().fit(X_train, y_train)
y_pred = reg.predict(X_test)
r2 = np.round(r2_score(y_test, y_pred), 2)
rmse = np.round(np.sqrt(mean_squared_error(y_test, y_pred)), 2)
end_time = time.time()
time_taken = np.round((end_time - start_time), 2)
temp_df.loc[len(temp_df)] = [dataset, dataform, X_train.shape[1], str(i).split('.')[-1][:-2], r2, rmse, time_taken]
return temp_dfOriginal data:
original_df = train_test_ML('AirQualityUCI', 'Original', X_train, y_train, X_test, y_test)
original_df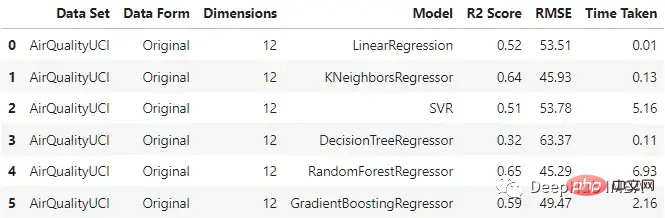
It can be seen that KNN regressor and random forest perform relatively well when inputting original data, and the training time of random forest is the longest.
PCA
pca_df = train_test_ML('AirQualityUCI', 'PCA Reduced', X_train_pca, y_train, X_test_pca, y_test)
pca_df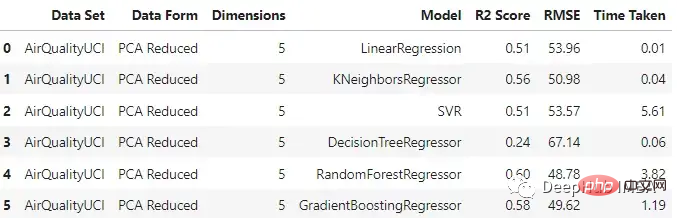
与原始数据集相比,不同模型的性能有不同程度的下降。梯度增强回归和支持向量回归在两种情况下保持了一致性。这里一个主要的差异也是预期的是模型训练所花费的时间。与其他模型不同的是,SVR在这两种情况下花费的时间差不多。
SVD
svd_df = train_test_ML('AirQualityUCI', 'SVD Reduced', X_train_svd, y_train, X_test_svd, y_test)
svd_df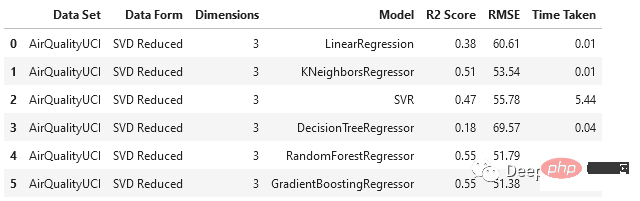
与PCA相比,SVD以更大的比例降低了维度,随机森林和梯度增强回归器的表现相对优于其他模型。
回归模型分析
对于这个数据集,使用主成分分析时,数据维数从12维降至5维,使用奇异值分析时,数据降至3维。
- 就机器学习性能而言,数据集的原始形式相对更好。造成这种情况的一个潜在原因可能是,当我们使用这种技术降低维数时,在这个过程中会发生信息损失。
- 但是线性回归、支持向量回归和梯度增强回归在原始和PCA案例中的表现是一致的。
- 在我们通过SVD得到的数据上,所有模型的性能都下降了。
- 在降维情况下,由于特征变量的维数较低,模型所花费的时间减少了。
将类似的过程应用于其他六个数据集进行测试,得到以下结果:
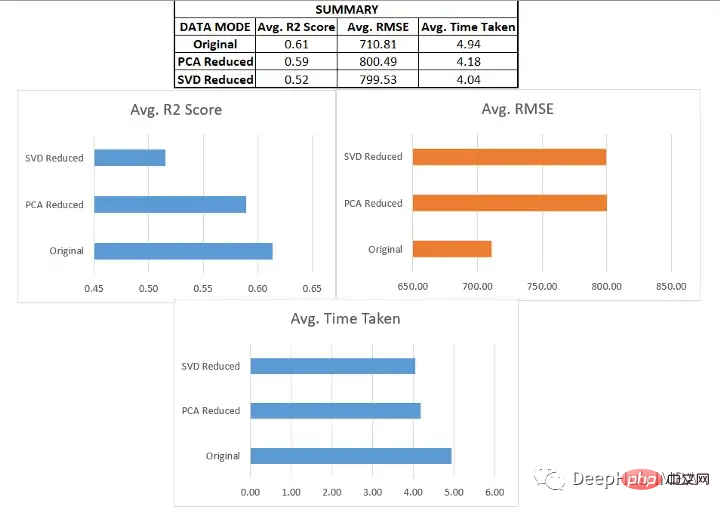
我们在各种数据集上使用了SVD和PCA,并对比了在原始高维特征空间上训练的回归模型与在约简特征空间上训练的模型的有效性
- 原始数据集始终优于由降维方法创建的低维数据。这说明在降维过程中可能丢失了一些信息。
- 当用于更大的数据集时,降维方法有助于显著减少数据集中的特征数量,从而提高机器学习模型的有效性。对于较小的数据集,改影响并不显著。
- 模型的性能在original和pca_reduced两种模式下保持一致。如果一个模型在原始数据集上表现得更好,那么它在PCA模式下也会表现得更好。同样,较差的模型也没有得到改进。
- 在SVD的情况下,模型的性能下降比较明显。这可能是n_components数量选择的问题,因为太小数量肯定会丢失数据。
- 决策树在SVD数据集时一直是非常差的,因为它本来就是一个弱学习器
训练分类模型
对于分类我们将使用另一种降维方法:LDA。机器学习和模式识别任务经常使用被称为线性判别分析(LDA)的降维方法。这种监督学习技术旨在最大化几个类或类别之间的距离,同时将数据投影到低维空间。由于它的作用是最大化类之间的差异,因此只能用于分类任务。
from sklearn.linear_model import LogisticRegression from sklearn.neighbors import KNeighborsClassifier from sklearn.svm import SVC from sklearn.tree import DecisionTreeClassifier from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier from sklearn.metrics import accuracy_score, f1_score, recall_score, precision_score
继续我们的训练方法
def train_test_ML2(dataset, dataform, X_train, y_train, X_test, y_test):
temp_df = pd.DataFrame(columns=['Data Set', 'Data Form', 'Dimensions', 'Model', 'Accuracy', 'F1 Score', 'Recall', 'Precision', 'Time Taken'])
for i in [LogisticRegression, KNeighborsClassifier, SVC, DecisionTreeClassifier, RandomForestClassifier, GradientBoostingClassifier]:
start_time = time.time()
reg = i().fit(X_train, y_train)
y_pred = reg.predict(X_test)
accuracy = np.round(accuracy_score(y_test, y_pred), 2)
f1 = np.round(f1_score(y_test, y_pred, average='weighted'), 2)
recall = np.round(recall_score(y_test, y_pred, average='weighted'), 2)
precision = np.round(precision_score(y_test, y_pred, average='weighted'), 2)
end_time = time.time()
time_taken = np.round((end_time - start_time), 2)
temp_df.loc[len(temp_df)] = [dataset, dataform, X_train.shape[1], str(i).split('.')[-1][:-2], accuracy, f1, recall, precision, time_taken]
return temp_df开始训练
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis lda = LinearDiscriminantAnalysis() X_train_lda = lda.fit_transform(X_train, y_train) X_test_lda = lda.transform(X_test)
预处理、分割和数据集的缩放,都与回归部分相同。在对8个不同的数据集进行新联后我们得到了下面结果:
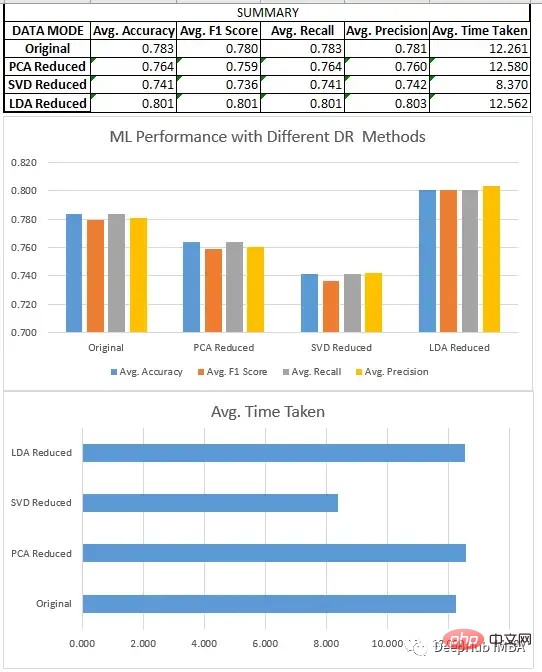
分类模型分析
我们比较了上面所有的三种方法SVD、LDA和PCA。
- LDA数据集通常优于原始形式的数据和由其他降维方法创建的低维数据,因为它旨在识别最有效区分类的特征的线性组合,而原始数据和其他无监督降维技术不关心数据集的标签。
- 降维技术在应用于更大的数据集时,可以极大地减少了数据集中的特征数量,这提高了机器学习模型的效率。在较小的数据集上,影响不是特别明显。除了LDA(它在这些情况下也很有效),因为它们在一些情况下,如二元分类,可以将数据集的维度减少到只有一个。
- 当我们在寻找一定的性能时,LDA可以是分类问题的一个非常好的起点。
- SVD与回归一样,模型的性能下降很明显。需要调整n_components的选择。
总结
我们比较了一些降维技术的性能,如奇异值分解(SVD)、主成分分析(PCA)和线性判别分析(LDA)。我们的研究结果表明,方法的选择取决于特定的数据集和手头的任务。
For regression tasks, we find that PCA generally performs better than SVD. In the case of classification, LDA outperforms SVD and PCA, as well as the original dataset. It is important that Linear Discriminant Analysis (LDA) consistently beats Principal Component Analysis (PCA) in classification tasks, but this does not mean that LDA is a better technique in general. This is because LDA is a supervised learning algorithm that relies on labeled data to locate the most discriminative features in the data, while PCA is an unsupervised technique that does not require labeled data and seeks to maintain as much variance as possible. Therefore, PCA may be better suited for unsupervised tasks or situations where interpretability is critical, while LDA may be better suited for tasks involving labeled data.
While dimensionality reduction techniques can help reduce the number of features in a dataset and improve the efficiency of machine learning models, it is important to consider the potential impact on model performance and result interpretability.
The complete code of this article:
https://github.com/salmankhi/DimensionalityReduction/blob/main/Notebook_25373.ipynb
The above is the detailed content of Comparison of common dimensionality reduction technologies: feasibility analysis of reducing data dimensions while maintaining information integrity. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

AI Hentai Generator
Generate AI Hentai for free.

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1378
1378
 52
52
 15 recommended open source free image annotation tools
Mar 28, 2024 pm 01:21 PM
15 recommended open source free image annotation tools
Mar 28, 2024 pm 01:21 PM
Image annotation is the process of associating labels or descriptive information with images to give deeper meaning and explanation to the image content. This process is critical to machine learning, which helps train vision models to more accurately identify individual elements in images. By adding annotations to images, the computer can understand the semantics and context behind the images, thereby improving the ability to understand and analyze the image content. Image annotation has a wide range of applications, covering many fields, such as computer vision, natural language processing, and graph vision models. It has a wide range of applications, such as assisting vehicles in identifying obstacles on the road, and helping in the detection and diagnosis of diseases through medical image recognition. . This article mainly recommends some better open source and free image annotation tools. 1.Makesens
 This article will take you to understand SHAP: model explanation for machine learning
Jun 01, 2024 am 10:58 AM
This article will take you to understand SHAP: model explanation for machine learning
Jun 01, 2024 am 10:58 AM
In the fields of machine learning and data science, model interpretability has always been a focus of researchers and practitioners. With the widespread application of complex models such as deep learning and ensemble methods, understanding the model's decision-making process has become particularly important. Explainable AI|XAI helps build trust and confidence in machine learning models by increasing the transparency of the model. Improving model transparency can be achieved through methods such as the widespread use of multiple complex models, as well as the decision-making processes used to explain the models. These methods include feature importance analysis, model prediction interval estimation, local interpretability algorithms, etc. Feature importance analysis can explain the decision-making process of a model by evaluating the degree of influence of the model on the input features. Model prediction interval estimate
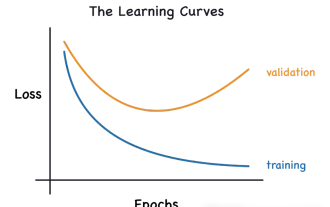 Identify overfitting and underfitting through learning curves
Apr 29, 2024 pm 06:50 PM
Identify overfitting and underfitting through learning curves
Apr 29, 2024 pm 06:50 PM
This article will introduce how to effectively identify overfitting and underfitting in machine learning models through learning curves. Underfitting and overfitting 1. Overfitting If a model is overtrained on the data so that it learns noise from it, then the model is said to be overfitting. An overfitted model learns every example so perfectly that it will misclassify an unseen/new example. For an overfitted model, we will get a perfect/near-perfect training set score and a terrible validation set/test score. Slightly modified: "Cause of overfitting: Use a complex model to solve a simple problem and extract noise from the data. Because a small data set as a training set may not represent the correct representation of all data." 2. Underfitting Heru
 Transparent! An in-depth analysis of the principles of major machine learning models!
Apr 12, 2024 pm 05:55 PM
Transparent! An in-depth analysis of the principles of major machine learning models!
Apr 12, 2024 pm 05:55 PM
In layman’s terms, a machine learning model is a mathematical function that maps input data to a predicted output. More specifically, a machine learning model is a mathematical function that adjusts model parameters by learning from training data to minimize the error between the predicted output and the true label. There are many models in machine learning, such as logistic regression models, decision tree models, support vector machine models, etc. Each model has its applicable data types and problem types. At the same time, there are many commonalities between different models, or there is a hidden path for model evolution. Taking the connectionist perceptron as an example, by increasing the number of hidden layers of the perceptron, we can transform it into a deep neural network. If a kernel function is added to the perceptron, it can be converted into an SVM. this one
 The evolution of artificial intelligence in space exploration and human settlement engineering
Apr 29, 2024 pm 03:25 PM
The evolution of artificial intelligence in space exploration and human settlement engineering
Apr 29, 2024 pm 03:25 PM
In the 1950s, artificial intelligence (AI) was born. That's when researchers discovered that machines could perform human-like tasks, such as thinking. Later, in the 1960s, the U.S. Department of Defense funded artificial intelligence and established laboratories for further development. Researchers are finding applications for artificial intelligence in many areas, such as space exploration and survival in extreme environments. Space exploration is the study of the universe, which covers the entire universe beyond the earth. Space is classified as an extreme environment because its conditions are different from those on Earth. To survive in space, many factors must be considered and precautions must be taken. Scientists and researchers believe that exploring space and understanding the current state of everything can help understand how the universe works and prepare for potential environmental crises
 Implementing Machine Learning Algorithms in C++: Common Challenges and Solutions
Jun 03, 2024 pm 01:25 PM
Implementing Machine Learning Algorithms in C++: Common Challenges and Solutions
Jun 03, 2024 pm 01:25 PM
Common challenges faced by machine learning algorithms in C++ include memory management, multi-threading, performance optimization, and maintainability. Solutions include using smart pointers, modern threading libraries, SIMD instructions and third-party libraries, as well as following coding style guidelines and using automation tools. Practical cases show how to use the Eigen library to implement linear regression algorithms, effectively manage memory and use high-performance matrix operations.
 Explainable AI: Explaining complex AI/ML models
Jun 03, 2024 pm 10:08 PM
Explainable AI: Explaining complex AI/ML models
Jun 03, 2024 pm 10:08 PM
Translator | Reviewed by Li Rui | Chonglou Artificial intelligence (AI) and machine learning (ML) models are becoming increasingly complex today, and the output produced by these models is a black box – unable to be explained to stakeholders. Explainable AI (XAI) aims to solve this problem by enabling stakeholders to understand how these models work, ensuring they understand how these models actually make decisions, and ensuring transparency in AI systems, Trust and accountability to address this issue. This article explores various explainable artificial intelligence (XAI) techniques to illustrate their underlying principles. Several reasons why explainable AI is crucial Trust and transparency: For AI systems to be widely accepted and trusted, users need to understand how decisions are made
 Five schools of machine learning you don't know about
Jun 05, 2024 pm 08:51 PM
Five schools of machine learning you don't know about
Jun 05, 2024 pm 08:51 PM
Machine learning is an important branch of artificial intelligence that gives computers the ability to learn from data and improve their capabilities without being explicitly programmed. Machine learning has a wide range of applications in various fields, from image recognition and natural language processing to recommendation systems and fraud detection, and it is changing the way we live. There are many different methods and theories in the field of machine learning, among which the five most influential methods are called the "Five Schools of Machine Learning". The five major schools are the symbolic school, the connectionist school, the evolutionary school, the Bayesian school and the analogy school. 1. Symbolism, also known as symbolism, emphasizes the use of symbols for logical reasoning and expression of knowledge. This school of thought believes that learning is a process of reverse deduction, through existing



