 Technology peripherals
Technology peripherals
 AI
AI
 Exploration and application of genome conformation prediction models and high-throughput computational genetic screening methods
Exploration and application of genome conformation prediction models and high-throughput computational genetic screening methods
Exploration and application of genome conformation prediction models and high-throughput computational genetic screening methods

Figure 0
#Differences in genome conformation in different types of cells determine the specificity of gene expression , thereby determining the functional differences of different cell types. For a long time, experimental methods for genome conformation detection, from in situ hybridization to high-throughput detection such as Hi-C and micro-C technologies, are usually time-consuming, labor-intensive, costly and have strong technical limitations. These methods greatly limit the widespread application of these experimental techniques in the field of genome conformation research, especially in the study of rare cell types and the need to verify the causal relationship of genome conformation regulation on a large scale. The limitations of these methods have also long restricted new discoveries in the field of three-dimensional genome conformational regulation.

##Picture 1
##January 9, 2023,NYU Grossman School of Medicine’s Aristotelis Tsirigos laboratory and Broad Institute of MIT and Harvard’s Xia Bo laboratory collaborated to publish an article in Nature Biotechnology "Cell type-specific prediction of 3D Chromatin organization enables high-throughput in silico genetic screening》.

In this study, the first authors Tan Jimin and Dr. Xia Bo, doctoral students at New York University School of Medicine, first proposed a new multi-modal machine learning model C.Origami. Predict the chromatin conformation of specific cell types and propose a new high-throughput computational genetic screening (ISGS) method
based on the principles of genetic screening to identify cell type-specific functions Genomic elements help discover new mechanisms of chromatin conformation regulation.
Figure 2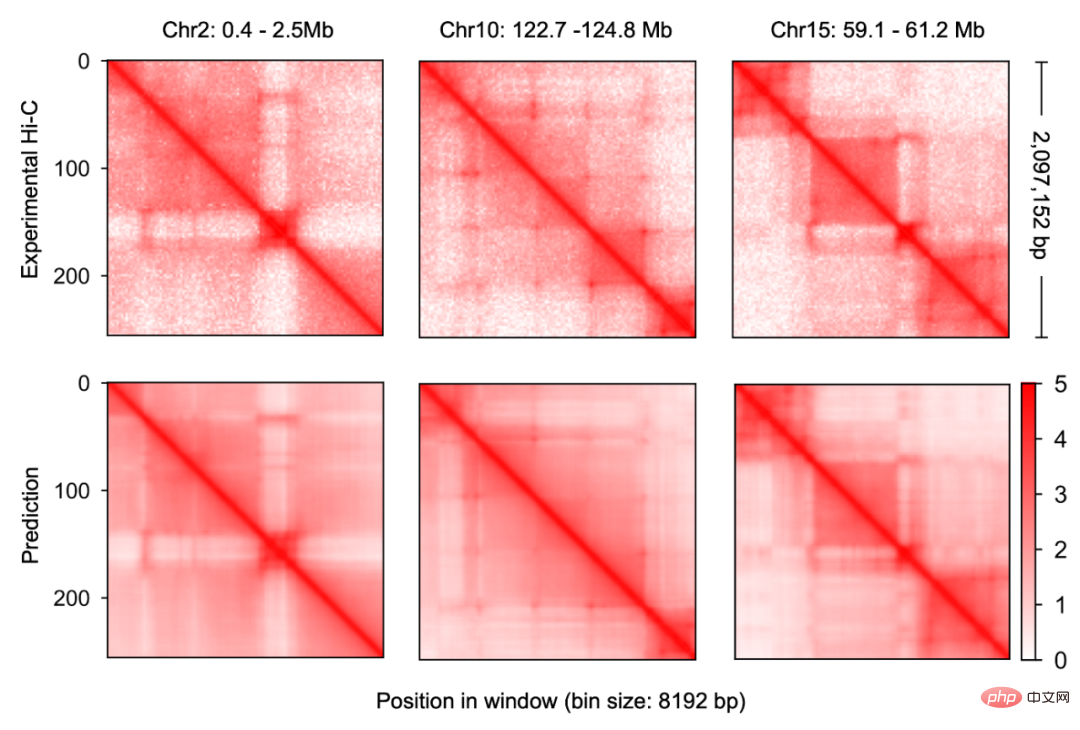
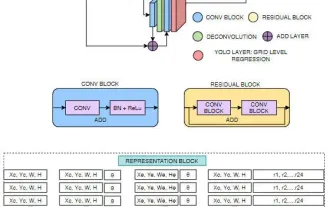
ResearcherFirst A new multi-modal deep learning framework, Origami
, was constructed for genomic data, enabling it to effectively integrate DNA sequence information and cell-specific functional genomic information to predict new genomic information. Through repeated debugging and model training, the researchers found that integrating DNA sequence, CTCF binding status (CTCF ChIP-seq), and ATAC-seq signals as input information can accurately predict chromatin conformation, and use the two-dimensional Hi-C matrix as Predict the output target (Figure 1-2). Input information was 2 million base pairs of DNA, CTCF ChIP-seq and ATAC-seq. Researchers use Onehot-encoding to encode discrete DNA sequences, while CTCF ChIP-seq and ATAC-seq encode non-discrete features. C.Origami model is divided into three parts, the encoder that processes and compresses DNA and genome information, the Transformer middle layer and the output Hi-C decoder. The encoder consists of a series of 1D ResNet and strided convolution to encode and compress the input information of 2 million base pairs. At the end of the encoder the 2 million length message is compressed to 256 length and used as the input message to the Transformer. Transformer's self-attention mechanism can handle the interdependency between different genomic regions and improve the overall performance of the model. The attention matrix in Transformer can also enhance the interpretability of the model. The researchers converted the attention weight into an "attention score" to measure the model's emphasis on different areas when predicting. Finally, the researchers converted the 1D output of the Transformer module into a 2D contact/adjacency matrix using "outer concatenation", which was used as input information for the Hi-C decoder. The decoder is a Dilated 2D ResNet. The researchers adjusted the dilation factors of different layers so that the receptive field at each pixel position of the final layer can cover all input information. This model for predicting chromatin conformation is called C.Origami. Researchers call C.Origami the first multimodal deep learning model in genomics. Due to its multimodal nature, C.Origami is able to accurately predict (de novo prediction) chromatin conformation in new cell types that have never been exposed before. For example, a model trained on IMR-90 cells (lung fibroblasts) was able to accurately predict specific chromatin conformations in GM12878 cells (B lymphocytes) (Figure 3). Figure 3 structural variant --- - Such as chromosomal translocations - are very common in tumors and often alter chromatin interaction patterns, which may affect the expression of oncogenes or tumor suppressor genes. Studying the effects of these structural variations on chromatin conformation and gene expression is important for understanding the mechanisms of tumor occurrence and progression. This type of research usually requires the use of experiments such as 4C-seq or Hi-C to analyze the chromatin conformation of structural variation sites, but is often limited by resources and time and is difficult to conduct on a large scale. In this study, C. Origami can model variations in DNA sequences in input variables and then predict new chromatin interactions in the mutated cancer genome. Previous studies found that the T-cell acute lymphoblastic leukemia (T-ALL) cell model CUTLL1 has a chr7-chr9 chromosomal translocation (Figure 4). By computationally simulating chromosomal translocation variants, C. Origami accurately predicted the new TAD structure at the variant site and detected a ‘chromatin stripe’ structure extending from chr9 to chr7 (Figure 4). Figure 4 In view of the accurate prediction effect of C.Origami, Inspired by the principle of reverse genetic screening, researchers proposed a new high-throughput computational genetic screening (ISGS) method to systematically identify cell type-specific functional genomic elements and help discover new ones. Staining regulatory molecules (Figure 5). The researchers developed a framework for computational genetic screening ISGS based on the C. Origami model for the systematic identification of cis-regulatory elements required for chromatin conformation. Using genome-wide 1kb resolution ISGS, the authors isolated cis-regulatory elements (∼1% of the genome) that have important effects on chromatin conformation. These chromatin conformation regulatory sequences exhibit differential dependence on CTCF binding and ATAC-seq signals (Fig. 5 ). Figure 5 The ISGS framework enables high-throughput screening of cell- or disease-specific chromatin conformations. The researchers performed ISGS in CUTLL1, Jurkat and normal T cells and found that a cis-regulatory element (CHD4-insu) near the CHD4 gene was specifically lost in T-ALL cells. Screening results indicate that the insulating loss of CHD4-insu in T-ALL cells may enable the CHD4 gene to establish new chromatin interactions, thereby upregulating CHD4 expression and promoting leukemia cell proliferation. ISGS can also be used to systematically discover novel trans-acting factors that regulate chromatin conformation. Through enrichment analysis of important cell type-specific regulatory sequences and transcription factor binding sites, the researchers identified regulatory factors that contribute to cell type-specific genome conformation. Interestingly, previous studies found that MAZ may regulate chromatin conformation together with CTCF. Through ISGS and transcription factor enrichment analysis, the authors found that MAZ is greatly enriched in open chromatin regions, while showing only weak binding in non-open chromatin regions where CTCF binds. This result suggests that MAZ may regulate genome conformation independently of CTCF. Researchers see great potential in multimodal machine learning models that combine DNA sequence and chromatin information in chromatin structure prediction. The underlying multimodal architecture of the model, Origami, can be extended to the application of other genomics data, such as epigenetic modifications, gene expression, functional screening of mutations, etc. Researchers predict that future genomics research will shift more towards the use of deep learning models as tools for primary computational genetic screening, supplemented by a new generation of high-throughput research methods validated by biological experiments. In this study, Tan Jimin, a doctoral candidate at New York University School of Medicine, is the first author, and Dr. Aristotelis Tsirigos and Dr. Xia Bo are the co-corresponding authors. This research began with the brainstorming of Xia Bo and Tan Jimin during the epidemic lockdown in October 2020. After two and a half years of improvement and polishing, it was officially published in Nature Biotechnology in January 2023. The code and training data of this project have been open sourced on GitHub and Zenodo, and are equipped with Google Colab for functional demonstration. Project address: https://github.com/tanjimin/C.Origami Dr. Xia Bo’s laboratory (Broad Institute of MIT and Harvard) homepage: www.boxialab.org Dr. Xia Bo is committed to analyzing the core mechanism and regulation of the three-dimensional conformation of the genome. Its biological significance for human disease, development and evolution. Xia Bo's laboratory welcomes like-minded postdocs to join the team. Tsirigos Lab (New York University Grossman School of Medicine) Home page: http://www.tsirigos.com Tsirigos Lab’s main page Research interests include the application of chromatin, epigenetics and machine learning in precision medicine. 


Corresponding author
The above is the detailed content of Exploration and application of genome conformation prediction models and high-throughput computational genetic screening methods. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

AI Hentai Generator
Generate AI Hentai for free.

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 The world's most powerful open source MoE model is here, with Chinese capabilities comparable to GPT-4, and the price is only nearly one percent of GPT-4-Turbo
May 07, 2024 pm 04:13 PM
The world's most powerful open source MoE model is here, with Chinese capabilities comparable to GPT-4, and the price is only nearly one percent of GPT-4-Turbo
May 07, 2024 pm 04:13 PM
Imagine an artificial intelligence model that not only has the ability to surpass traditional computing, but also achieves more efficient performance at a lower cost. This is not science fiction, DeepSeek-V2[1], the world’s most powerful open source MoE model is here. DeepSeek-V2 is a powerful mixture of experts (MoE) language model with the characteristics of economical training and efficient inference. It consists of 236B parameters, 21B of which are used to activate each marker. Compared with DeepSeek67B, DeepSeek-V2 has stronger performance, while saving 42.5% of training costs, reducing KV cache by 93.3%, and increasing the maximum generation throughput to 5.76 times. DeepSeek is a company exploring general artificial intelligence
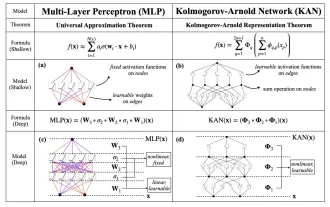 KAN, which replaces MLP, has been extended to convolution by open source projects
Jun 01, 2024 pm 10:03 PM
KAN, which replaces MLP, has been extended to convolution by open source projects
Jun 01, 2024 pm 10:03 PM
Earlier this month, researchers from MIT and other institutions proposed a very promising alternative to MLP - KAN. KAN outperforms MLP in terms of accuracy and interpretability. And it can outperform MLP running with a larger number of parameters with a very small number of parameters. For example, the authors stated that they used KAN to reproduce DeepMind's results with a smaller network and a higher degree of automation. Specifically, DeepMind's MLP has about 300,000 parameters, while KAN only has about 200 parameters. KAN has a strong mathematical foundation like MLP. MLP is based on the universal approximation theorem, while KAN is based on the Kolmogorov-Arnold representation theorem. As shown in the figure below, KAN has
 Hello, electric Atlas! Boston Dynamics robot comes back to life, 180-degree weird moves scare Musk
Apr 18, 2024 pm 07:58 PM
Hello, electric Atlas! Boston Dynamics robot comes back to life, 180-degree weird moves scare Musk
Apr 18, 2024 pm 07:58 PM
Boston Dynamics Atlas officially enters the era of electric robots! Yesterday, the hydraulic Atlas just "tearfully" withdrew from the stage of history. Today, Boston Dynamics announced that the electric Atlas is on the job. It seems that in the field of commercial humanoid robots, Boston Dynamics is determined to compete with Tesla. After the new video was released, it had already been viewed by more than one million people in just ten hours. The old people leave and new roles appear. This is a historical necessity. There is no doubt that this year is the explosive year of humanoid robots. Netizens commented: The advancement of robots has made this year's opening ceremony look like a human, and the degree of freedom is far greater than that of humans. But is this really not a horror movie? At the beginning of the video, Atlas is lying calmly on the ground, seemingly on his back. What follows is jaw-dropping
 Google is ecstatic: JAX performance surpasses Pytorch and TensorFlow! It may become the fastest choice for GPU inference training
Apr 01, 2024 pm 07:46 PM
Google is ecstatic: JAX performance surpasses Pytorch and TensorFlow! It may become the fastest choice for GPU inference training
Apr 01, 2024 pm 07:46 PM
The performance of JAX, promoted by Google, has surpassed that of Pytorch and TensorFlow in recent benchmark tests, ranking first in 7 indicators. And the test was not done on the TPU with the best JAX performance. Although among developers, Pytorch is still more popular than Tensorflow. But in the future, perhaps more large models will be trained and run based on the JAX platform. Models Recently, the Keras team benchmarked three backends (TensorFlow, JAX, PyTorch) with the native PyTorch implementation and Keras2 with TensorFlow. First, they select a set of mainstream
 AI subverts mathematical research! Fields Medal winner and Chinese-American mathematician led 11 top-ranked papers | Liked by Terence Tao
Apr 09, 2024 am 11:52 AM
AI subverts mathematical research! Fields Medal winner and Chinese-American mathematician led 11 top-ranked papers | Liked by Terence Tao
Apr 09, 2024 am 11:52 AM
AI is indeed changing mathematics. Recently, Tao Zhexuan, who has been paying close attention to this issue, forwarded the latest issue of "Bulletin of the American Mathematical Society" (Bulletin of the American Mathematical Society). Focusing on the topic "Will machines change mathematics?", many mathematicians expressed their opinions. The whole process was full of sparks, hardcore and exciting. The author has a strong lineup, including Fields Medal winner Akshay Venkatesh, Chinese mathematician Zheng Lejun, NYU computer scientist Ernest Davis and many other well-known scholars in the industry. The world of AI has changed dramatically. You know, many of these articles were submitted a year ago.
 Time Series Forecasting NLP Large Model New Work: Automatically Generate Implicit Prompts for Time Series Forecasting
Mar 18, 2024 am 09:20 AM
Time Series Forecasting NLP Large Model New Work: Automatically Generate Implicit Prompts for Time Series Forecasting
Mar 18, 2024 am 09:20 AM
Today I would like to share a recent research work from the University of Connecticut that proposes a method to align time series data with large natural language processing (NLP) models on the latent space to improve the performance of time series forecasting. The key to this method is to use latent spatial hints (prompts) to enhance the accuracy of time series predictions. Paper title: S2IP-LLM: SemanticSpaceInformedPromptLearningwithLLMforTimeSeriesForecasting Download address: https://arxiv.org/pdf/2403.05798v1.pdf 1. Large problem background model
 Tesla robots work in factories, Musk: The degree of freedom of hands will reach 22 this year!
May 06, 2024 pm 04:13 PM
Tesla robots work in factories, Musk: The degree of freedom of hands will reach 22 this year!
May 06, 2024 pm 04:13 PM
The latest video of Tesla's robot Optimus is released, and it can already work in the factory. At normal speed, it sorts batteries (Tesla's 4680 batteries) like this: The official also released what it looks like at 20x speed - on a small "workstation", picking and picking and picking: This time it is released One of the highlights of the video is that Optimus completes this work in the factory, completely autonomously, without human intervention throughout the process. And from the perspective of Optimus, it can also pick up and place the crooked battery, focusing on automatic error correction: Regarding Optimus's hand, NVIDIA scientist Jim Fan gave a high evaluation: Optimus's hand is the world's five-fingered robot. One of the most dexterous. Its hands are not only tactile
 FisheyeDetNet: the first target detection algorithm based on fisheye camera
Apr 26, 2024 am 11:37 AM
FisheyeDetNet: the first target detection algorithm based on fisheye camera
Apr 26, 2024 am 11:37 AM
Target detection is a relatively mature problem in autonomous driving systems, among which pedestrian detection is one of the earliest algorithms to be deployed. Very comprehensive research has been carried out in most papers. However, distance perception using fisheye cameras for surround view is relatively less studied. Due to large radial distortion, standard bounding box representation is difficult to implement in fisheye cameras. To alleviate the above description, we explore extended bounding box, ellipse, and general polygon designs into polar/angular representations and define an instance segmentation mIOU metric to analyze these representations. The proposed model fisheyeDetNet with polygonal shape outperforms other models and simultaneously achieves 49.5% mAP on the Valeo fisheye camera dataset for autonomous driving





