 Technology peripherals
Technology peripherals
 AI
AI
 Using AI to explore the 'key” to antibodies and accelerate drug development—an interview with the Baitu Biotechnology team
Using AI to explore the 'key” to antibodies and accelerate drug development—an interview with the Baitu Biotechnology team
Using AI to explore the 'key” to antibodies and accelerate drug development—an interview with the Baitu Biotechnology team
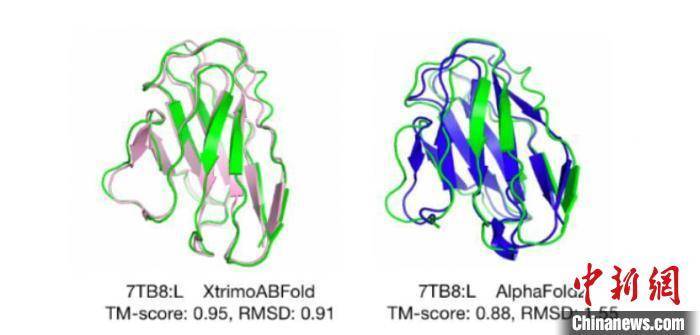
Baitu Biotechnology provides a picture comparing the performance of "AlphaFold2" and "xTrimoABFold" antibody structure prediction
China News Service, Beijing, June 12th, Title: Using AI to explore the "key" to antibodies and accelerate drug research and development - an interview with the Baitu Biotechnology team
Reporter Zhang Su
Just imagine, the proteins present in the human body have various forms of "keyholes". If researchers can accurately predict the forms and design the corresponding "keys", they can greatly improve the "unlocking" efficiency, and this has Help accelerate more cutting-edge drug research and development and other life science projects...
The reporter learned from the interview that the R&D team of Baitu Biotechnology has made a series of positive progress, including the release of the AIGP (AI Generated Protein) platform driven by large life science models.
Collaborative research and development, innovative ideas and methods for drug development
In the recent period, Chen Zibo's laboratory at West Lake University and Baitu Biotech jointly carried out an AND-gate protein design research project based on the AIGP platform. This move aims to design artificial "smart" proteins with molecular logic gate functions to achieve precise signal response and regulation of complex living systems and provide innovative solutions for the diagnosis and treatment of many diseases.
We use the AIGP platform to quickly design and generate proteins with specific properties. This provides a basis for the design and application of AND-gate proteins, and also provides a new way of thinking and methods for the development of innovative drugs. "Liu Wei, CEO of Baitu Biotech, said in an interview.

The picture shows Liu Wei, CEO of Baitu Biotechnology. Photo provided by Baitu Biotechnology
Liu Wei also cited the example of cooperation with Logos Biotechnology to explore new tumor targets, using the self-developed tumor immune escape target identification platform (Tier-A) to quickly transform innovative results into new mechanisms of action. For biological macromolecules, "It usually takes several months to analyze the crystal structure in the laboratory, but the AIGP platform shortens this process to one or two weeks."
According to Song Le, chief technology officer and chief AI scientist of Baitu Biotechnology, target discovery, compound synthesis and screening are key links in the research and development of new drugs, and large AI models are expected to achieve double improvements in the efficiency and effectiveness of the process, " We hope to quickly design a 'key' that meets the 'keyhole' precision in the computer, speed up the evolution of artificially designed proteins, and thus solve the pain points in the life sciences industry."
It is reported that Baitu Biotech will soon disclose part of the user interface of the AIGP platform to potential partners and professional researchers. They plan to launch AIGP 2.0 version within a year to provide more mission model training and other capabilities.
Faster and more accurate, promoting model iteration and application implementation
The AIGP platform with its many highlights relies on the large-scale multi-modal model system "xTrimo".
It is reported that “xTrimo” is composed of a pre-trained model with hundreds of billions of parameters and multiple downstream task models. Its model adopts the design logic of a four-layer nested structure. In April this year, Baitu Biotech released data showing that the antibody structure prediction model "xTrimoABFold" showed higher accuracy and efficiency. Compared with "AlphaFold2", its inference speed is 540 times faster, its running speed is 151 times faster, and it can accurately predict the atomic-level structure of antibodies within 3 seconds.
“The accuracy or quality of data will have a direct impact on model performance.” Song Le said when explaining the reasons for obtaining this “report card”. The R&D team also conducted detailed alignment work on public data and supplemented high-quality data with internal laboratory data to effectively address the challenges of data mining and utilization. The biological supercomputing platform they established before can dynamically obtain thousands to tens of thousands of GPUs and corresponding CPU resources, which is very powerful in terms of computing power.
"xTrimoGene", another sub-model in the "xTrimo" system, and its version "scFoundation" for more downstream tasks have also received widespread attention. Recently, the results of a paper released by Baitu Biotechnology and Tsinghua University show that xTrimoGene and scFoundation outperform classic methods in six basic tasks and downstream application scenarios in the single-cell field, including cell type annotation, perturbation prediction, and synergistic drug combination. Realized industry SOTA.
Our goal is to do the best in basic work and further improve the accuracy and efficiency of the model, including increasing the speed of antibody structure prediction to milliseconds. " Song Le said.
At the same time, Liu Wei said that they also hope to open some capabilities to the industry, recruit more ecological partners, promote the rapid iteration and application of large models, and form a high-quality data closed loop.
Facing the future and creating more and better proteins
First, scientific researchers input various parameter requirements for the target protein, and the algorithm generates the protein. Then, the protein is automatically printed out, and the researchers only need to take the protein automatically produced by the algorithm for further scientific verification... This is Liu Wei's vision for the future "protein factory".
Looking forward to pushing the AIGP platform to a new height through our efforts in the next five years, bringing more beautiful proteins to the world, and contributing to creating a better life. "He said.
Baitu Biotech has launched the "Excellent Developer Program", which is open to cutting-edge biotechnology experts, drug development experts and clinical professional teams, and aims to target the future. Song Le explained that the plan will provide scientific research funding and technical support for high-level translational medical research projects. He said that Baitu Biotechnology has established a team composed of talents with multi-disciplinary backgrounds to ensure that scientific research results continue to be produced.
It should be noted that as the application field of AI protein design continues to expand, the cooperation carried out by relevant parties based on the AIGP platform is not limited to the development of antibody drugs in a narrow sense, but also enters a variety of macromolecule drugs, cell gene therapy, life sciences Tools and other fields, and extends to environmental protection, petroleum, food and other fields, using AI's ability to generate innovative proteins to solve more real-life problems. For example, researchers are studying proteases in an effort to find a way to efficiently break down plastics or speed up preparations for specific energy production.
Liu Wei said that while working hard to expand target areas and business models, they maintain the same original intention, which is to “use AI large models, cutting-edge biocomputing technology, etc. to model complex life forms and design new proteins to meet the challenge.” Despite the huge investment, they will still move forward firmly on this path. (over)
The above is the detailed content of Using AI to explore the 'key” to antibodies and accelerate drug development—an interview with the Baitu Biotechnology team. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

AI Hentai Generator
Generate AI Hentai for free.

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1378
1378
 52
52
 How to implement file sorting by debian readdir
Apr 13, 2025 am 09:06 AM
How to implement file sorting by debian readdir
Apr 13, 2025 am 09:06 AM
In Debian systems, the readdir function is used to read directory contents, but the order in which it returns is not predefined. To sort files in a directory, you need to read all files first, and then sort them using the qsort function. The following code demonstrates how to sort directory files using readdir and qsort in Debian system: #include#include#include#include#include//Custom comparison function, used for qsortintcompare(constvoid*a,constvoid*b){returnstrcmp(*(
 How to optimize the performance of debian readdir
Apr 13, 2025 am 08:48 AM
How to optimize the performance of debian readdir
Apr 13, 2025 am 08:48 AM
In Debian systems, readdir system calls are used to read directory contents. If its performance is not good, try the following optimization strategy: Simplify the number of directory files: Split large directories into multiple small directories as much as possible, reducing the number of items processed per readdir call. Enable directory content caching: build a cache mechanism, update the cache regularly or when directory content changes, and reduce frequent calls to readdir. Memory caches (such as Memcached or Redis) or local caches (such as files or databases) can be considered. Adopt efficient data structure: If you implement directory traversal by yourself, select more efficient data structures (such as hash tables instead of linear search) to store and access directory information
 How debian readdir integrates with other tools
Apr 13, 2025 am 09:42 AM
How debian readdir integrates with other tools
Apr 13, 2025 am 09:42 AM
The readdir function in the Debian system is a system call used to read directory contents and is often used in C programming. This article will explain how to integrate readdir with other tools to enhance its functionality. Method 1: Combining C language program and pipeline First, write a C program to call the readdir function and output the result: #include#include#include#includeintmain(intargc,char*argv[]){DIR*dir;structdirent*entry;if(argc!=2){
 Debian mail server firewall configuration tips
Apr 13, 2025 am 11:42 AM
Debian mail server firewall configuration tips
Apr 13, 2025 am 11:42 AM
Configuring a Debian mail server's firewall is an important step in ensuring server security. The following are several commonly used firewall configuration methods, including the use of iptables and firewalld. Use iptables to configure firewall to install iptables (if not already installed): sudoapt-getupdatesudoapt-getinstalliptablesView current iptables rules: sudoiptables-L configuration
 How to configure firewall rules for Debian syslog
Apr 13, 2025 am 06:51 AM
How to configure firewall rules for Debian syslog
Apr 13, 2025 am 06:51 AM
This article describes how to configure firewall rules using iptables or ufw in Debian systems and use Syslog to record firewall activities. Method 1: Use iptablesiptables is a powerful command line firewall tool in Debian system. View existing rules: Use the following command to view the current iptables rules: sudoiptables-L-n-v allows specific IP access: For example, allow IP address 192.168.1.100 to access port 80: sudoiptables-AINPUT-ptcp--dport80-s192.16
 How to set the Debian Apache log level
Apr 13, 2025 am 08:33 AM
How to set the Debian Apache log level
Apr 13, 2025 am 08:33 AM
This article describes how to adjust the logging level of the ApacheWeb server in the Debian system. By modifying the configuration file, you can control the verbose level of log information recorded by Apache. Method 1: Modify the main configuration file to locate the configuration file: The configuration file of Apache2.x is usually located in the /etc/apache2/ directory. The file name may be apache2.conf or httpd.conf, depending on your installation method. Edit configuration file: Open configuration file with root permissions using a text editor (such as nano): sudonano/etc/apache2/apache2.conf
 How to learn Debian syslog
Apr 13, 2025 am 11:51 AM
How to learn Debian syslog
Apr 13, 2025 am 11:51 AM
This guide will guide you to learn how to use Syslog in Debian systems. Syslog is a key service in Linux systems for logging system and application log messages. It helps administrators monitor and analyze system activity to quickly identify and resolve problems. 1. Basic knowledge of Syslog The core functions of Syslog include: centrally collecting and managing log messages; supporting multiple log output formats and target locations (such as files or networks); providing real-time log viewing and filtering functions. 2. Install and configure Syslog (using Rsyslog) The Debian system uses Rsyslog by default. You can install it with the following command: sudoaptupdatesud
 How Debian OpenSSL prevents man-in-the-middle attacks
Apr 13, 2025 am 10:30 AM
How Debian OpenSSL prevents man-in-the-middle attacks
Apr 13, 2025 am 10:30 AM
In Debian systems, OpenSSL is an important library for encryption, decryption and certificate management. To prevent a man-in-the-middle attack (MITM), the following measures can be taken: Use HTTPS: Ensure that all network requests use the HTTPS protocol instead of HTTP. HTTPS uses TLS (Transport Layer Security Protocol) to encrypt communication data to ensure that the data is not stolen or tampered during transmission. Verify server certificate: Manually verify the server certificate on the client to ensure it is trustworthy. The server can be manually verified through the delegate method of URLSession



