The importance of cross-validation cannot be ignored!
In order not to change the original meaning, what needs to be re-expressed is: First, we need to figure out why cross-validation is needed?
Cross-validation is a technique commonly used in machine learning and statistics to evaluate the performance and generalization ability of a predictive model, especially when data are limited or to evaluate the model's generalization to new unseen data. Cross-validation is extremely valuable when it comes to capabilities.

Under what circumstances will cross-validation be used?
- Model performance evaluation: Cross-validation helps estimate the performance of the model on unseen data. By training and evaluating the model on multiple subsets of the data, cross-validation provides a more robust estimate of model performance than a single train-test split.
- Data efficiency: When data is limited, cross-validation makes full use of all available samples, providing a more reliable assessment of model performance by using all data simultaneously for training and evaluation.
- Hyperparameter tuning: Cross validation is often used to select the best hyperparameters for a model. By evaluating the performance of your model using different hyperparameter settings on different subsets of the data, you can identify the hyperparameter values that perform best in terms of overall performance.
- Detecting overfitting: Cross-validation helps detect whether the model is overfitting the training data. If the model performs significantly better on the training set than on the validation set, it may indicate overfitting and requires adjustments, such as regularization or choosing a simpler model.
- Generalization ability evaluation: Cross-validation provides an evaluation of the model's ability to generalize to unseen data. By evaluating the model on multiple data splits, it helps evaluate the model's ability to capture underlying patterns in the data without relying on randomness or a specific train-test split.
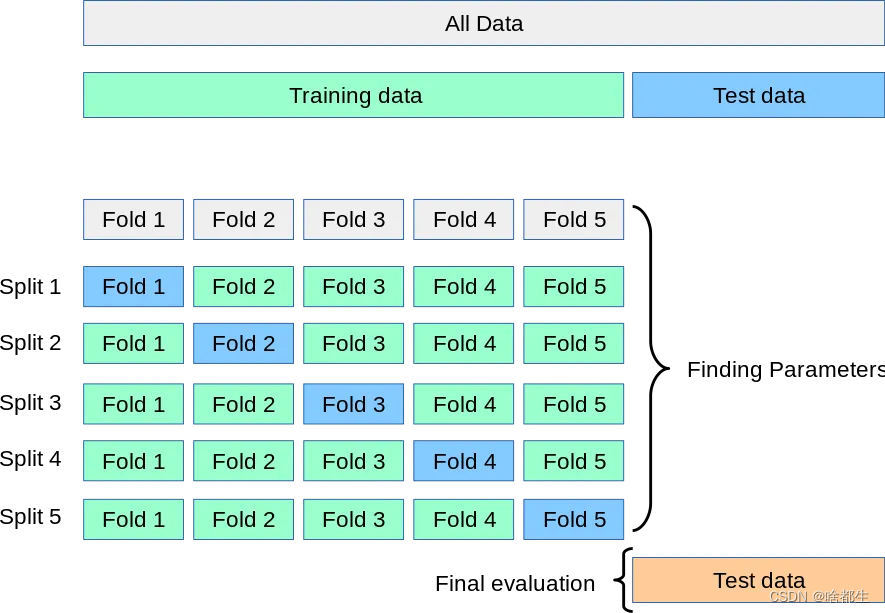
The general idea of cross-validation can be shown in Figure 5 fold cross. In each iteration, the new model is trained on four sub-datasets and performed on the last retained sub-dataset. Test to make sure all data is utilized. Through indicators such as average score and standard deviation, a true measure of model performance is provided.
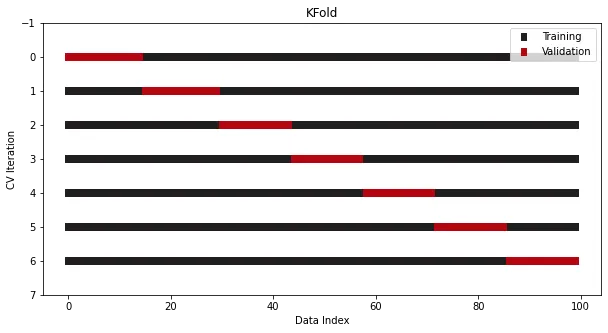
Everything has to start with K-fold crossover.
KFold
K-fold cross-validation has been integrated in Sklearn. Here is a 7-fold example:
from sklearn.datasets import make_regressionfrom sklearn.model_selection import KFoldx, y = make_regression(n_samples=100)# Init the splittercross_validation = KFold(n_splits=7)
There is another A common operation is to perform a Shuffle before performing the split, which further minimizes the risk of overfitting by destroying the original order of the samples:
cross_validation = KFold(n_splits=7, shuffle=True)
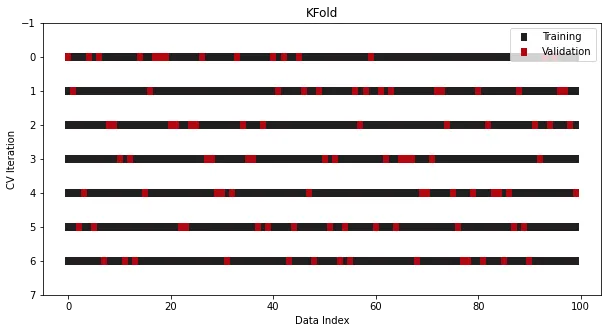
In this way, a simple k-fold Cross-validation can be done, be sure to check the source code! Be sure to check out the source code! Be sure to check out the source code!
StratifiedKFold
StratifiedKFold is specially designed for classification problems.
In some classification problems, the target distribution should remain unchanged even if the data is divided into multiple sets. For example, in most cases, a binary target with a class ratio of 30 to 70 should still maintain the same ratio in the training set and the test set. In ordinary KFold, this rule is broken because the data is shuffled before splitting. , the category proportions will not be maintained.
To solve this problem, another splitter class specifically for classification is used in Sklearn - StratifiedKFold:
from sklearn.datasets import make_classificationfrom sklearn.model_selection import StratifiedKFoldx, y = make_classification(n_samples=100, n_classes=2)cross_validation = StratifiedKFold(n_splits=7, shuffle=True, random_state=1121218)
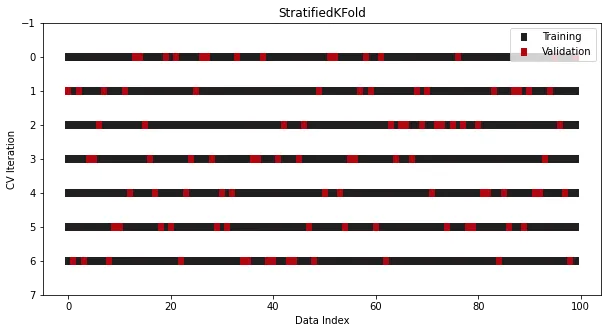
Although it is different from KFold Looks similar, but now the class proportions remain consistent across all splits and iterations Cross-validation is very similar
Logically speaking, by using different random seeds to generate multiple training/test sets, it should be similar to a robust cross-validation process in enough iterations
Scikit-learn library also provides corresponding interfaces:
from sklearn.model_selection import ShuffleSplitcross_validation = ShuffleSplit(n_splits=7, train_size=0.75, test_size=0.25)
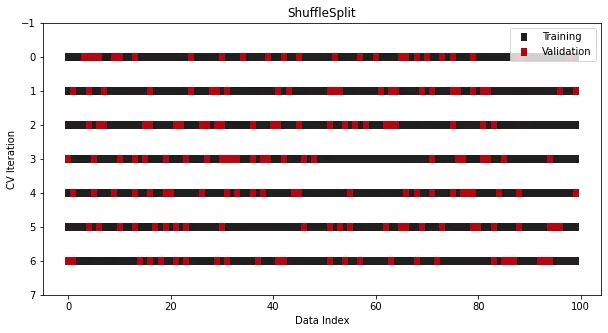 When the data set is a time series, traditional cross-validation cannot be used , this will completely disrupt the order. In order to solve this problem, refer to Sklearn provides another splitter-TimeSeriesSplit,
When the data set is a time series, traditional cross-validation cannot be used , this will completely disrupt the order. In order to solve this problem, refer to Sklearn provides another splitter-TimeSeriesSplit,
from sklearn.model_selection import TimeSeriesSplitcross_validation = TimeSeriesSplit(n_splits=7)
Cross-validation of non-independent and identically distributed (non-IID) data
The above method is processed for independent and identically distributed data sets, that is, the process of generating data will not be affected by other samples
However, in some cases, the data does not meet the conditions of independent and identical distribution (IID), that is, there is a dependency between some samples. This situation also occurs in Kaggle competitions, such as the Google Brain Ventilator Pressure competition. This data records the air pressure values of the artificial lung during thousands of breaths (inhalation and exhalation), and is recorded at every moment of each breath. There are approximately 80 rows of data for each breathing process, and these rows are related to each other. In this case, traditional cross-validation methods cannot be used because the division of the data may "happen just in the middle of a breathing process"
This can be understood as the need to "group" these data, Because the data within the group are related. For example, when collecting medical data from multiple patients, each patient has multiple samples. However, these data are likely to be affected by individual patient differences and therefore also need to be grouped
Often we hope that a model trained on a specific group will generalize well to other unseen groups. groups, so when performing cross-validation, "tag" the data of these groups and tell them how to distinguish them from each other.
Several interfaces are provided in Sklearn to handle these situations:
- GroupKFold
- StratifiedGroupKFold
- LeaveOneGroupOut
- LeavePGroupsOut
- GroupShuffleSplit
It is strongly recommended to understand the idea of cross-validation and how to implement it. It is a good way to look at the Sklearn source code. In addition, you need to have a clear definition of your own data set, and data preprocessing is really important.
The above is the detailed content of The importance of cross-validation cannot be ignored!. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

AI Hentai Generator
Generate AI Hentai for free.

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1377
1377
 52
52
 15 recommended open source free image annotation tools
Mar 28, 2024 pm 01:21 PM
15 recommended open source free image annotation tools
Mar 28, 2024 pm 01:21 PM
Image annotation is the process of associating labels or descriptive information with images to give deeper meaning and explanation to the image content. This process is critical to machine learning, which helps train vision models to more accurately identify individual elements in images. By adding annotations to images, the computer can understand the semantics and context behind the images, thereby improving the ability to understand and analyze the image content. Image annotation has a wide range of applications, covering many fields, such as computer vision, natural language processing, and graph vision models. It has a wide range of applications, such as assisting vehicles in identifying obstacles on the road, and helping in the detection and diagnosis of diseases through medical image recognition. . This article mainly recommends some better open source and free image annotation tools. 1.Makesens
 This article will take you to understand SHAP: model explanation for machine learning
Jun 01, 2024 am 10:58 AM
This article will take you to understand SHAP: model explanation for machine learning
Jun 01, 2024 am 10:58 AM
In the fields of machine learning and data science, model interpretability has always been a focus of researchers and practitioners. With the widespread application of complex models such as deep learning and ensemble methods, understanding the model's decision-making process has become particularly important. Explainable AI|XAI helps build trust and confidence in machine learning models by increasing the transparency of the model. Improving model transparency can be achieved through methods such as the widespread use of multiple complex models, as well as the decision-making processes used to explain the models. These methods include feature importance analysis, model prediction interval estimation, local interpretability algorithms, etc. Feature importance analysis can explain the decision-making process of a model by evaluating the degree of influence of the model on the input features. Model prediction interval estimate
 Transparent! An in-depth analysis of the principles of major machine learning models!
Apr 12, 2024 pm 05:55 PM
Transparent! An in-depth analysis of the principles of major machine learning models!
Apr 12, 2024 pm 05:55 PM
In layman’s terms, a machine learning model is a mathematical function that maps input data to a predicted output. More specifically, a machine learning model is a mathematical function that adjusts model parameters by learning from training data to minimize the error between the predicted output and the true label. There are many models in machine learning, such as logistic regression models, decision tree models, support vector machine models, etc. Each model has its applicable data types and problem types. At the same time, there are many commonalities between different models, or there is a hidden path for model evolution. Taking the connectionist perceptron as an example, by increasing the number of hidden layers of the perceptron, we can transform it into a deep neural network. If a kernel function is added to the perceptron, it can be converted into an SVM. this one
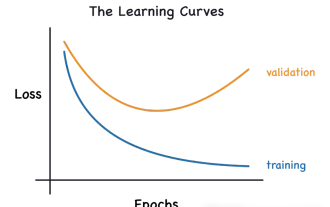 Identify overfitting and underfitting through learning curves
Apr 29, 2024 pm 06:50 PM
Identify overfitting and underfitting through learning curves
Apr 29, 2024 pm 06:50 PM
This article will introduce how to effectively identify overfitting and underfitting in machine learning models through learning curves. Underfitting and overfitting 1. Overfitting If a model is overtrained on the data so that it learns noise from it, then the model is said to be overfitting. An overfitted model learns every example so perfectly that it will misclassify an unseen/new example. For an overfitted model, we will get a perfect/near-perfect training set score and a terrible validation set/test score. Slightly modified: "Cause of overfitting: Use a complex model to solve a simple problem and extract noise from the data. Because a small data set as a training set may not represent the correct representation of all data." 2. Underfitting Heru
 The evolution of artificial intelligence in space exploration and human settlement engineering
Apr 29, 2024 pm 03:25 PM
The evolution of artificial intelligence in space exploration and human settlement engineering
Apr 29, 2024 pm 03:25 PM
In the 1950s, artificial intelligence (AI) was born. That's when researchers discovered that machines could perform human-like tasks, such as thinking. Later, in the 1960s, the U.S. Department of Defense funded artificial intelligence and established laboratories for further development. Researchers are finding applications for artificial intelligence in many areas, such as space exploration and survival in extreme environments. Space exploration is the study of the universe, which covers the entire universe beyond the earth. Space is classified as an extreme environment because its conditions are different from those on Earth. To survive in space, many factors must be considered and precautions must be taken. Scientists and researchers believe that exploring space and understanding the current state of everything can help understand how the universe works and prepare for potential environmental crises
 Implementing Machine Learning Algorithms in C++: Common Challenges and Solutions
Jun 03, 2024 pm 01:25 PM
Implementing Machine Learning Algorithms in C++: Common Challenges and Solutions
Jun 03, 2024 pm 01:25 PM
Common challenges faced by machine learning algorithms in C++ include memory management, multi-threading, performance optimization, and maintainability. Solutions include using smart pointers, modern threading libraries, SIMD instructions and third-party libraries, as well as following coding style guidelines and using automation tools. Practical cases show how to use the Eigen library to implement linear regression algorithms, effectively manage memory and use high-performance matrix operations.
 Explainable AI: Explaining complex AI/ML models
Jun 03, 2024 pm 10:08 PM
Explainable AI: Explaining complex AI/ML models
Jun 03, 2024 pm 10:08 PM
Translator | Reviewed by Li Rui | Chonglou Artificial intelligence (AI) and machine learning (ML) models are becoming increasingly complex today, and the output produced by these models is a black box – unable to be explained to stakeholders. Explainable AI (XAI) aims to solve this problem by enabling stakeholders to understand how these models work, ensuring they understand how these models actually make decisions, and ensuring transparency in AI systems, Trust and accountability to address this issue. This article explores various explainable artificial intelligence (XAI) techniques to illustrate their underlying principles. Several reasons why explainable AI is crucial Trust and transparency: For AI systems to be widely accepted and trusted, users need to understand how decisions are made
 Is Flash Attention stable? Meta and Harvard found that their model weight deviations fluctuated by orders of magnitude
May 30, 2024 pm 01:24 PM
Is Flash Attention stable? Meta and Harvard found that their model weight deviations fluctuated by orders of magnitude
May 30, 2024 pm 01:24 PM
MetaFAIR teamed up with Harvard to provide a new research framework for optimizing the data bias generated when large-scale machine learning is performed. It is known that the training of large language models often takes months and uses hundreds or even thousands of GPUs. Taking the LLaMA270B model as an example, its training requires a total of 1,720,320 GPU hours. Training large models presents unique systemic challenges due to the scale and complexity of these workloads. Recently, many institutions have reported instability in the training process when training SOTA generative AI models. They usually appear in the form of loss spikes. For example, Google's PaLM model experienced up to 20 loss spikes during the training process. Numerical bias is the root cause of this training inaccuracy,




