 Technology peripherals
Technology peripherals
 AI
AI
 NVIDIA, Mila, and Caltech jointly release multi-modal molecular structure-text model of LLM combined with drug discovery
NVIDIA, Mila, and Caltech jointly release multi-modal molecular structure-text model of LLM combined with drug discovery
NVIDIA, Mila, and Caltech jointly release multi-modal molecular structure-text model of LLM combined with drug discovery

Author | Liu Shengchao
Editor | Kaixia
Starting in 2021, the combination of big language and multi-modality has swept the machine learning research community .
With the development of large models and multi-modal applications, can we apply these techniques to drug discovery? And, can these natural language textual descriptions bring new perspectives to this challenging problem? The answer is yes, and we are optimistic about it
Recently, Canada’s Montreal Institute for Learning Algorithms (Mila), NVIDIA Research, University of Illinois at Urbana-Champaign (UIUC), Princeton University and California Institute of Technology The college's research team jointly learns the chemical structure and text description of molecules through comparative learning strategies, and proposes a multi-modal molecular structure-text model MoleculeSTM.
The research is titled "Multi-modal molecule structure–text model for text-based retrieval and editing" and was published in "Nature Machine Intelligence" on December 18, 2023.

Paper link: https://www.nature.com/articles/s42256-023-00759-6 needs to be rewritten
Among them, Dr. Liu Shengchao is the first author, and Professor Anima Anandkumar of NVIDIA Research is the corresponding author. Nie Weili, Wang Chengpeng, Lu Jiarui, Qiao Zhuoran, Liu Ling, Tang Jian and Xiao Chaowei are co-authors.
This project was conducted by Dr. Liu Shengchao after joining NVIDIA Research in March 2022, under the guidance of Teachers Nie Weili, Teacher Tang Jian, Teacher Xiao Chaowei and Teacher Anima Anandkumar.
Dr. Liu Shengchao said: "Our motivation was to conduct preliminary exploration of LLM and drug discovery, and finally proposed MoleculeSTM."
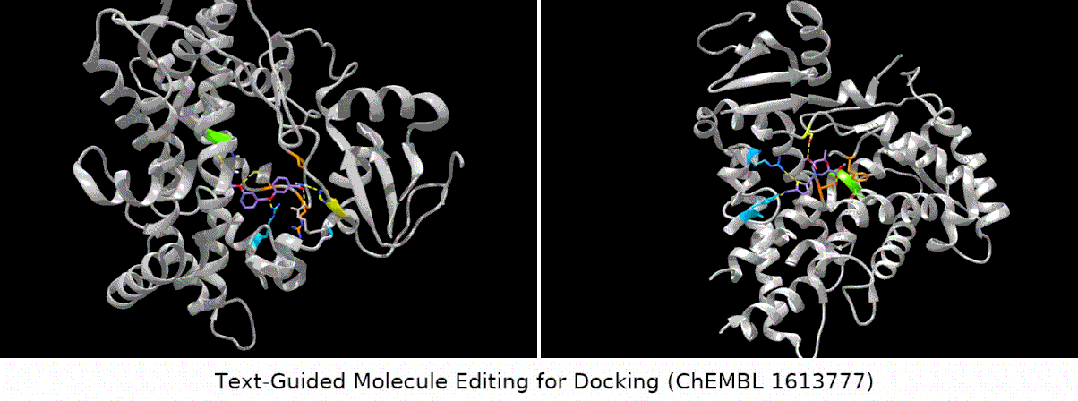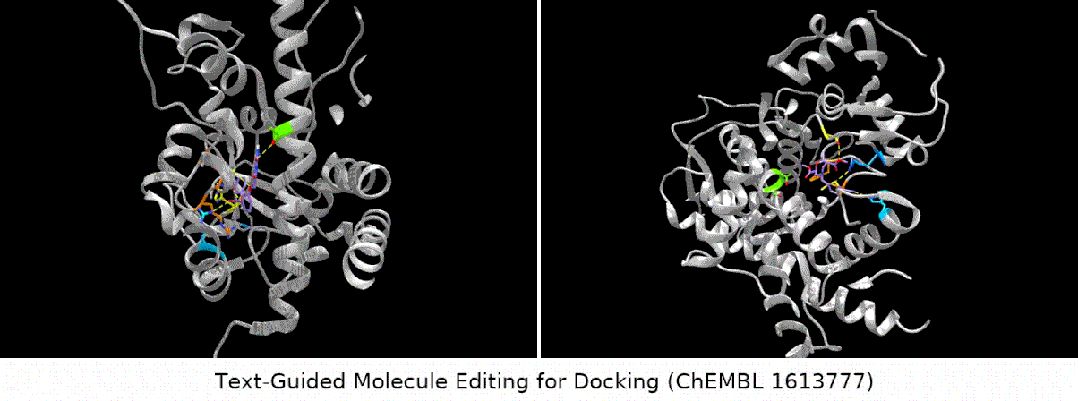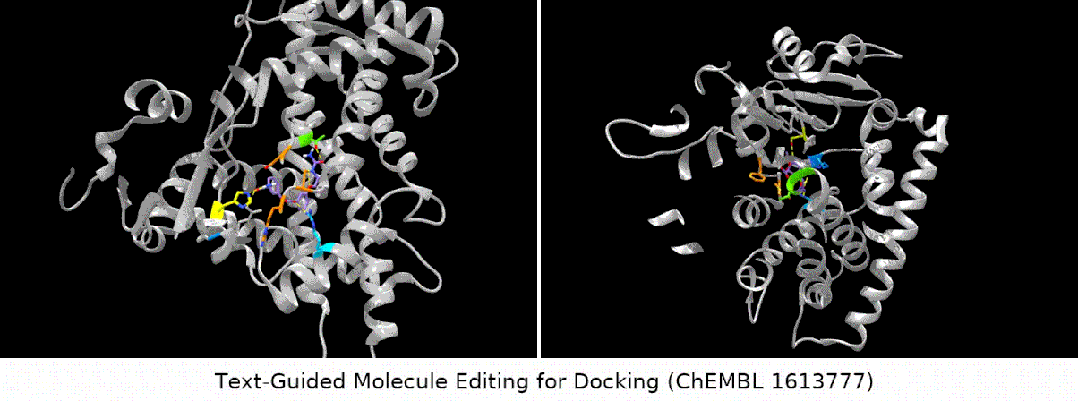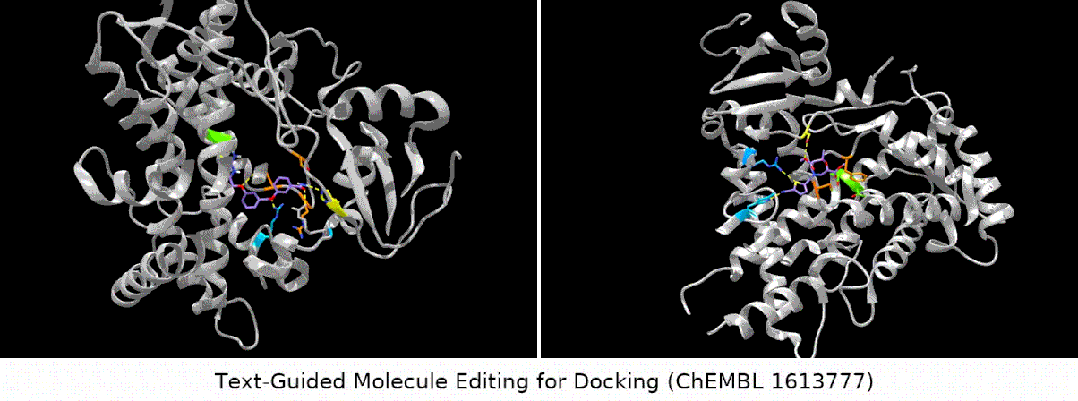
For docking The text is designed to guide molecule editing
The core idea of MoleculeSTM is very simple and direct, that is, the description of molecules can be divided into two categories: internal chemical structure and external function description. Here we use a contrastive pre-training method to align and connect these two types of information. The specific diagram is shown in the figure below
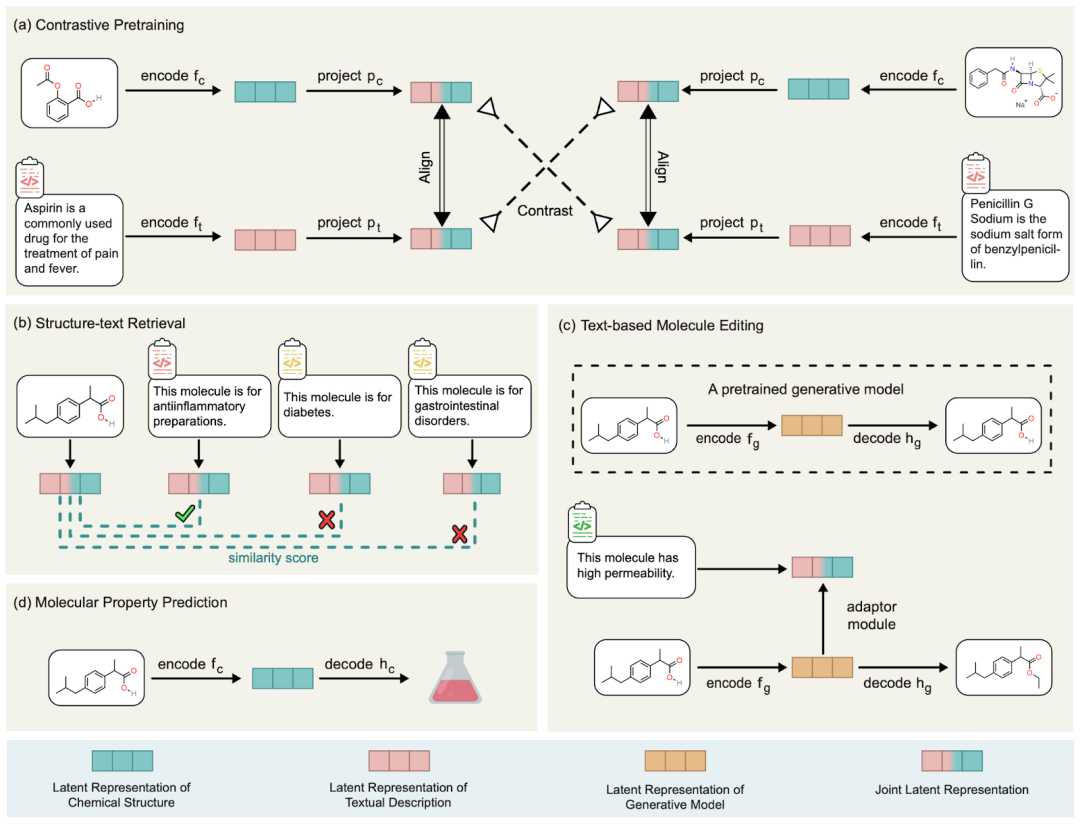
#Illustration: MoleculeSTM flow chart.
This alignment of MoleculeSTM has a very good property: when there are some tasks that are difficult to solve in the chemical space, we can transfer them to the natural language space. And natural language tasks will be relatively easier to solve due to its characteristics. Based on this, we designed a wide variety of downstream tasks to verify its effectiveness. Below we discuss several insights in detail.
Features of Natural Language and Large Language Models
In MoleculeSTM, we pose a problem for the first time. We take advantage of the open vocabulary and combinatorial characteristics of natural language
- Open vocabulary means that we can express all current human knowledge in natural language, so new knowledge that will appear in the future can also be expressed in modern language. Some language is used to summarize and summarize. For example, if a new protein appears, we hope to describe its function in natural language.
- Compositionality means that in natural language, a complex concept can be jointly expressed by several simple concepts. This is very helpful for tasks such as multi-attribute editing: it is very difficult to edit molecules to meet multiple properties at the same time in chemical space, but we can express multiple properties very easily in natural language.
In our recent work ChatDrug (https://arxiv.org/abs/2305.18090), we explored the conversational properties between natural language and large language models, which are of interest to Friends can go and take a look
Task design derived from features refers to the design of planning and arranging tasks based on the characteristics of the product or system
For existing language- For image tasks, they can be viewed as art-related tasks, such as generating pictures or text. That is, their results are varied and uncertain. However, scientific discoveries are scientific problems that usually have relatively clear results, such as the generation of small molecules with specific functions. This brings greater challenges in task design
In MoleculeSTM (Appendix B), we proposed two guidelines:
- The first task we consider is to be able to perform calculations and simulations to obtain results. In the future, wet-lab verification results will be considered, but this is not within the scope of the current work.
- Secondly, we only consider problems with ambiguous results. Specific examples include making a certain molecule more water-soluble or penetrable. Some problems have clear results, such as adding a certain functional group to a certain position in a molecule. We believe that such tasks are simpler and more straightforward for drug and chemistry experts. So it can be used as a proof-of-concept task in the future, but it will not become the main task target.
From this we designed three broad categories of tasks:
- Zero-shot structured text retrieval;
- Zero-shot text-based molecule editing ;
- Molecular property prediction.
We will focus on the second task in the following sections
The qualitative results of molecular editing are restated as follows:
This The task is to input a molecule and a natural language description (such as additional attributes) at the same time, and then hope to output a new molecule described in a composite language text. This is text-guided lead optimization.
The specific method is to use the already trained molecule generation model and our pre-trained MoleculeSTM to learn the alignment of the two latent spaces (latent space), thereby performing latent space interpolation, and then generate it through decoding target molecule. The process diagram is as follows.
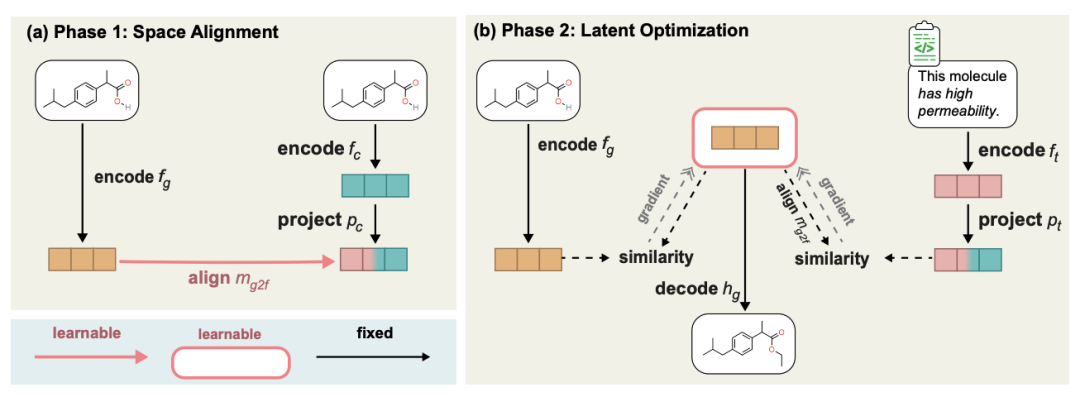
The content that needs to be rewritten is: a two-stage process diagram of zero-sample text-guided molecule editing
Here we show several groups of molecule editing The qualitative results are restated as follows: (Result details of the remaining downstream tasks can be found in the original paper). We mainly consider four types of molecular editing tasks:
- Single attribute editing: Editing a single attribute, such as water solubility, penetrability, and the number of hydrogen bond donors and acceptors.
- Composite attribute editing: Edit multiple attributes at the same time, such as water solubility and the number of hydrogen bond donors.
- Drug similarity editor: (Appendix D.5) makes the input molecule and the target molecule drug look closer.
- Neighbor search for patented drugs: For drugs that have been patented, drugs in the process are often reported together. Here we are combining the intermediate drug with the natural language description to see if it can generate the final target drug.
- binding affinity editor: We selected several ChEMBL assays as targets, with the goal of having higher binding affinity between the input molecules and the targets.
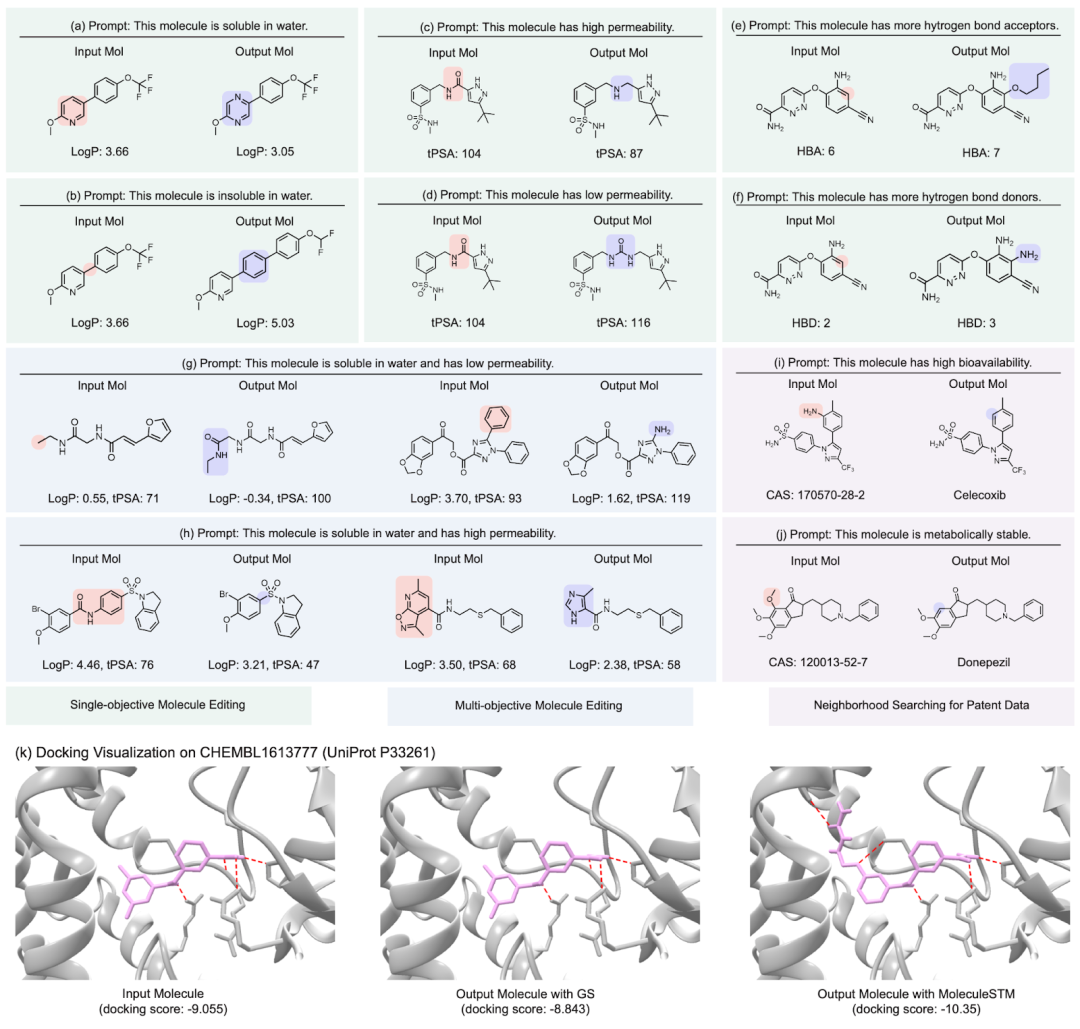
#Result display: zero-sample text-guided molecule editing. (Note: This is a direct translation of the original sentence into Chinese.)
What is more interesting is the last type of task. We found that MoleculeSTM can indeed perform matching based on the text description of the target protein. Optimization of lead compounds for ligands. (Note: The protein structure information here will only be known after evaluation.)
The above is the detailed content of NVIDIA, Mila, and Caltech jointly release multi-modal molecular structure-text model of LLM combined with drug discovery. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

AI Hentai Generator
Generate AI Hentai for free.

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1378
1378
 52
52
 Breaking through the boundaries of traditional defect detection, 'Defect Spectrum' achieves ultra-high-precision and rich semantic industrial defect detection for the first time.
Jul 26, 2024 pm 05:38 PM
Breaking through the boundaries of traditional defect detection, 'Defect Spectrum' achieves ultra-high-precision and rich semantic industrial defect detection for the first time.
Jul 26, 2024 pm 05:38 PM
In modern manufacturing, accurate defect detection is not only the key to ensuring product quality, but also the core of improving production efficiency. However, existing defect detection datasets often lack the accuracy and semantic richness required for practical applications, resulting in models unable to identify specific defect categories or locations. In order to solve this problem, a top research team composed of Hong Kong University of Science and Technology Guangzhou and Simou Technology innovatively developed the "DefectSpectrum" data set, which provides detailed and semantically rich large-scale annotation of industrial defects. As shown in Table 1, compared with other industrial data sets, the "DefectSpectrum" data set provides the most defect annotations (5438 defect samples) and the most detailed defect classification (125 defect categories
 NVIDIA dialogue model ChatQA has evolved to version 2.0, with the context length mentioned at 128K
Jul 26, 2024 am 08:40 AM
NVIDIA dialogue model ChatQA has evolved to version 2.0, with the context length mentioned at 128K
Jul 26, 2024 am 08:40 AM
The open LLM community is an era when a hundred flowers bloom and compete. You can see Llama-3-70B-Instruct, QWen2-72B-Instruct, Nemotron-4-340B-Instruct, Mixtral-8x22BInstruct-v0.1 and many other excellent performers. Model. However, compared with proprietary large models represented by GPT-4-Turbo, open models still have significant gaps in many fields. In addition to general models, some open models that specialize in key areas have been developed, such as DeepSeek-Coder-V2 for programming and mathematics, and InternVL for visual-language tasks.
 Google AI won the IMO Mathematical Olympiad silver medal, the mathematical reasoning model AlphaProof was launched, and reinforcement learning is so back
Jul 26, 2024 pm 02:40 PM
Google AI won the IMO Mathematical Olympiad silver medal, the mathematical reasoning model AlphaProof was launched, and reinforcement learning is so back
Jul 26, 2024 pm 02:40 PM
For AI, Mathematical Olympiad is no longer a problem. On Thursday, Google DeepMind's artificial intelligence completed a feat: using AI to solve the real question of this year's International Mathematical Olympiad IMO, and it was just one step away from winning the gold medal. The IMO competition that just ended last week had six questions involving algebra, combinatorics, geometry and number theory. The hybrid AI system proposed by Google got four questions right and scored 28 points, reaching the silver medal level. Earlier this month, UCLA tenured professor Terence Tao had just promoted the AI Mathematical Olympiad (AIMO Progress Award) with a million-dollar prize. Unexpectedly, the level of AI problem solving had improved to this level before July. Do the questions simultaneously on IMO. The most difficult thing to do correctly is IMO, which has the longest history, the largest scale, and the most negative
 Training with millions of crystal data to solve the crystallographic phase problem, the deep learning method PhAI is published in Science
Aug 08, 2024 pm 09:22 PM
Training with millions of crystal data to solve the crystallographic phase problem, the deep learning method PhAI is published in Science
Aug 08, 2024 pm 09:22 PM
Editor |KX To this day, the structural detail and precision determined by crystallography, from simple metals to large membrane proteins, are unmatched by any other method. However, the biggest challenge, the so-called phase problem, remains retrieving phase information from experimentally determined amplitudes. Researchers at the University of Copenhagen in Denmark have developed a deep learning method called PhAI to solve crystal phase problems. A deep learning neural network trained using millions of artificial crystal structures and their corresponding synthetic diffraction data can generate accurate electron density maps. The study shows that this deep learning-based ab initio structural solution method can solve the phase problem at a resolution of only 2 Angstroms, which is equivalent to only 10% to 20% of the data available at atomic resolution, while traditional ab initio Calculation
 Nature's point of view: The testing of artificial intelligence in medicine is in chaos. What should be done?
Aug 22, 2024 pm 04:37 PM
Nature's point of view: The testing of artificial intelligence in medicine is in chaos. What should be done?
Aug 22, 2024 pm 04:37 PM
Editor | ScienceAI Based on limited clinical data, hundreds of medical algorithms have been approved. Scientists are debating who should test the tools and how best to do so. Devin Singh witnessed a pediatric patient in the emergency room suffer cardiac arrest while waiting for treatment for a long time, which prompted him to explore the application of AI to shorten wait times. Using triage data from SickKids emergency rooms, Singh and colleagues built a series of AI models that provide potential diagnoses and recommend tests. One study showed that these models can speed up doctor visits by 22.3%, speeding up the processing of results by nearly 3 hours per patient requiring a medical test. However, the success of artificial intelligence algorithms in research only verifies this
 To provide a new scientific and complex question answering benchmark and evaluation system for large models, UNSW, Argonne, University of Chicago and other institutions jointly launched the SciQAG framework
Jul 25, 2024 am 06:42 AM
To provide a new scientific and complex question answering benchmark and evaluation system for large models, UNSW, Argonne, University of Chicago and other institutions jointly launched the SciQAG framework
Jul 25, 2024 am 06:42 AM
Editor |ScienceAI Question Answering (QA) data set plays a vital role in promoting natural language processing (NLP) research. High-quality QA data sets can not only be used to fine-tune models, but also effectively evaluate the capabilities of large language models (LLM), especially the ability to understand and reason about scientific knowledge. Although there are currently many scientific QA data sets covering medicine, chemistry, biology and other fields, these data sets still have some shortcomings. First, the data form is relatively simple, most of which are multiple-choice questions. They are easy to evaluate, but limit the model's answer selection range and cannot fully test the model's ability to answer scientific questions. In contrast, open-ended Q&A
 PRO | Why are large models based on MoE more worthy of attention?
Aug 07, 2024 pm 07:08 PM
PRO | Why are large models based on MoE more worthy of attention?
Aug 07, 2024 pm 07:08 PM
In 2023, almost every field of AI is evolving at an unprecedented speed. At the same time, AI is constantly pushing the technological boundaries of key tracks such as embodied intelligence and autonomous driving. Under the multi-modal trend, will the situation of Transformer as the mainstream architecture of AI large models be shaken? Why has exploring large models based on MoE (Mixed of Experts) architecture become a new trend in the industry? Can Large Vision Models (LVM) become a new breakthrough in general vision? ...From the 2023 PRO member newsletter of this site released in the past six months, we have selected 10 special interpretations that provide in-depth analysis of technological trends and industrial changes in the above fields to help you achieve your goals in the new year. be prepared. This interpretation comes from Week50 2023
 Automatically identify the best molecules and reduce synthesis costs. MIT develops a molecular design decision-making algorithm framework
Jun 22, 2024 am 06:43 AM
Automatically identify the best molecules and reduce synthesis costs. MIT develops a molecular design decision-making algorithm framework
Jun 22, 2024 am 06:43 AM
Editor | Ziluo AI’s use in streamlining drug discovery is exploding. Screen billions of candidate molecules for those that may have properties needed to develop new drugs. There are so many variables to consider, from material prices to the risk of error, that weighing the costs of synthesizing the best candidate molecules is no easy task, even if scientists use AI. Here, MIT researchers developed SPARROW, a quantitative decision-making algorithm framework, to automatically identify the best molecular candidates, thereby minimizing synthesis costs while maximizing the likelihood that the candidates have the desired properties. The algorithm also determined the materials and experimental steps needed to synthesize these molecules. SPARROW takes into account the cost of synthesizing a batch of molecules at once, since multiple candidate molecules are often available



