In drug development, it is crucial to determine the binding affinity and functional effect of small molecule ligands on proteins. Current computational methods can predict these protein-ligand interaction properties, but without high-resolution protein structures, accuracy is often lost and functional effects cannot be predicted.
Researchers at Monash University and Griffith University have developed PSICHIC (PhySIcoCHhemICal Graph Neural Network), a framework that combines physicochemical constraints directly from sequences Data decoding interaction fingerprints. This enables PSICHIC to decode the mechanisms behind protein-ligand interactions, achieving state-of-the-art accuracy and interpretability.
Trained on the same protein-ligand pairs without structural data, PSICHIC performed on par with, or even exceeded, leading structure-based methods in binding affinity predictions.
PSICHIC’s interpretable fingerprint identifies protein residues and ligand atoms involved in the interaction and helps reveal the selectivity determinants of protein-ligand interactions.
The research was titled "Physicochemical graph neural network for learning protein–ligand interaction fingerprints from sequence data" and was published in "Nature Machine Intelligence" on June 17, 2024.

In the drug discovery process, it is critical to understand the binding affinity and functional effects of small molecule ligands on proteins, as the selective interaction of the ligand with a specific protein determines The expected effect of the drug.
However, although current computational methods are capable of predicting protein-ligand interaction properties, in the absence of high-resolution protein structures, the prediction accuracy often decreases, and there are also difficulties in predicting functional effects.
Although sequence-based methods have more advantages in cost and resources (for example, they do not require expensive experimental structure determination processes), these methods often face the problem of excessive degrees of freedom in pattern matching, which can easily lead to overfitting and limited generalization. ization capabilities, thereby creating a performance gap with structure- or composite-based methods.
Physical Chemistry Graph Neural Network
A research team from Monash University and Griffith University developed PSICHIC (Physical Chemistry Graph Neural Network), a method to directly decode protein-ligands from sequence data following physical and chemical principles. Body interaction fingerprint method. Unlike previous sequence-based models, PSICHIC uniquely incorporates physicochemical constraints to achieve state-of-the-art accuracy and interpretability.
As a 2D sequence-based method, PSICHIC generates and imposes these constraints on a 2D plot by applying a clustering algorithm, allowing PSICHIC to primarily adapt to the rational underlying patterns that determine protein-ligand interactions during training.

(Source: Paper)
Performance Validation and Comparison
After training on the same protein-ligand pairs without structural data, PSICHIC outperforms in binding affinity predictions State-of-the-art structure-based and composite-based methods rival or even surpass them.
Experimental results on PDBBind v2016 and PDBBind v2020 datasets show that PSICHIC outperforms other sequence-based methods, such as TransCPI, MolTrans, and DrugBAN, on multiple indicators.
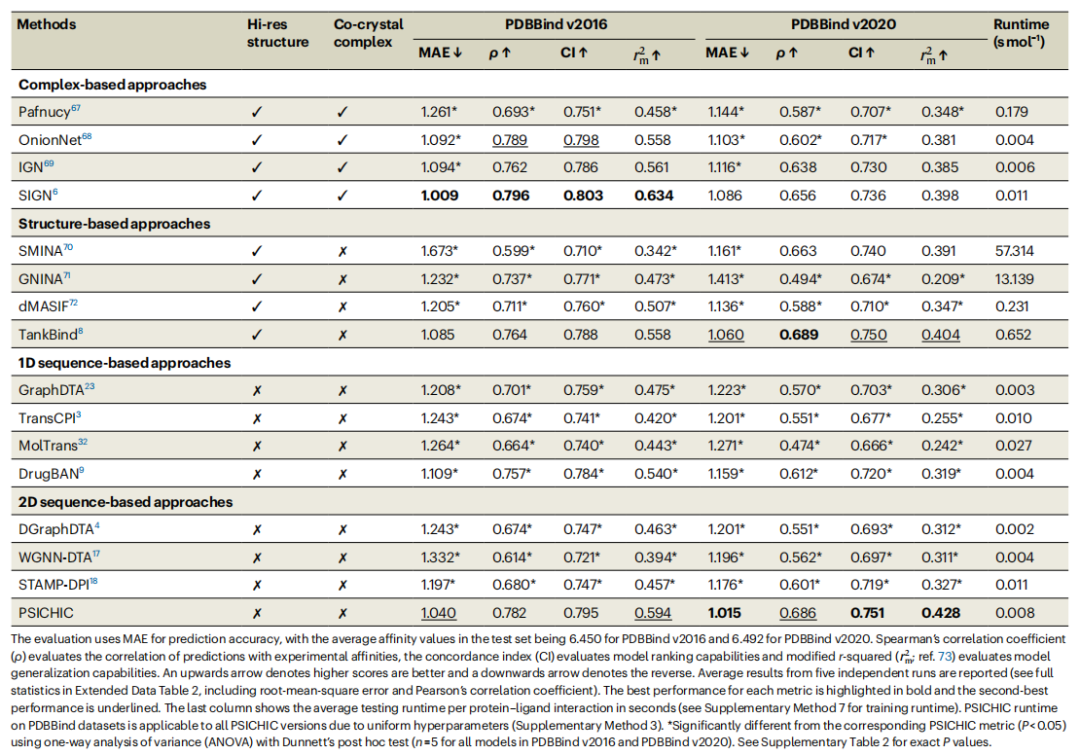
Performance statistical summary of protein-ligand binding affinity predictions on PDBBind v2016 and PDBBind v2020 benchmarks. (Source: paper)
Specifically:
Also:
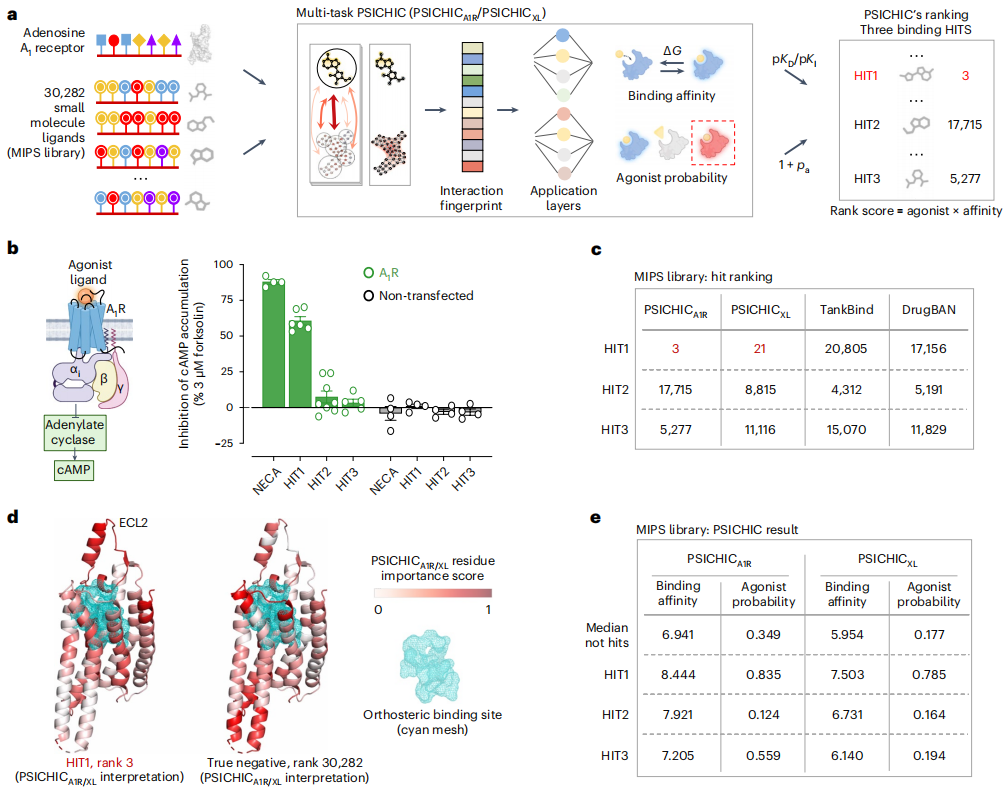
有趣的是,PSICHIC 的可解释指纹表明,它获得了仅从序列数据解码蛋白质-配体相互作用的潜在机制、识别结合位点蛋白质残基和所涉及的配体原子的能力,即使仅在具有结合亲和力标签而没有相互作用信息的序列数据上进行训练也是如此。
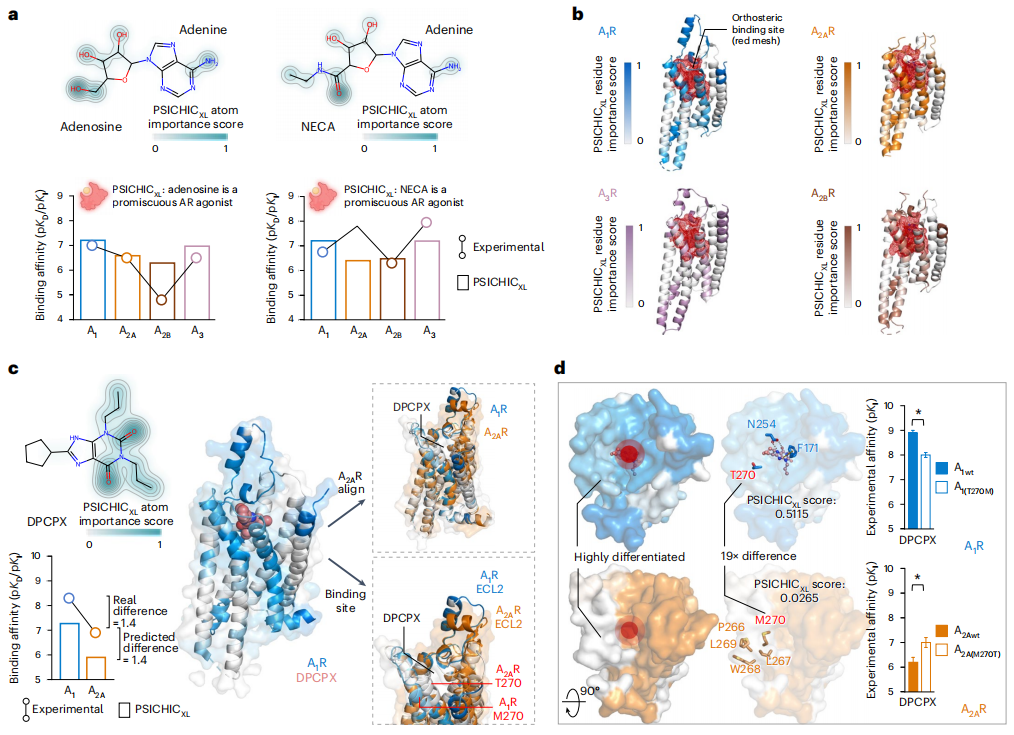
未来展望
以上是Nature子刊,准确率达96%,AI从序列中预测蛋白-配体互作的详细内容。更多信息请关注PHP中文网其他相关文章!




